There is a running joke in the 1990’s sitcom Friends that no one quite understands what Chandler Bing does for a living. They know it is “something to do with computers” but beyond that nothing. It is a little like that in my household. I see my wife’s eyes glazing over as I try and describe what I do.
There is a running joke in the 1990’s sitcom Friends that no one quite understands what Chandler Bing does for a living. They know it is “something to do with computers” but beyond that nothing. It is a little like that in my household. I see my wife’s eyes glazing over as I try and describe what I do.
This week I have a good example of spending time on a small cog that makes a bigger machine work and I think I can explain why it is interesting.
Our herbarium contains around three million specimens and we have a team of people busily databasing and imaging them so that they are accessible via the internet. Part of this process is that we submit data about the specimens to multiple databases such as the Global Biodiversity Information Facility (GBIF) and Global Plants section of JSTOR (an academic publications repository). Doing this means that researchers can see our data in the context of data from other institutions around the world.
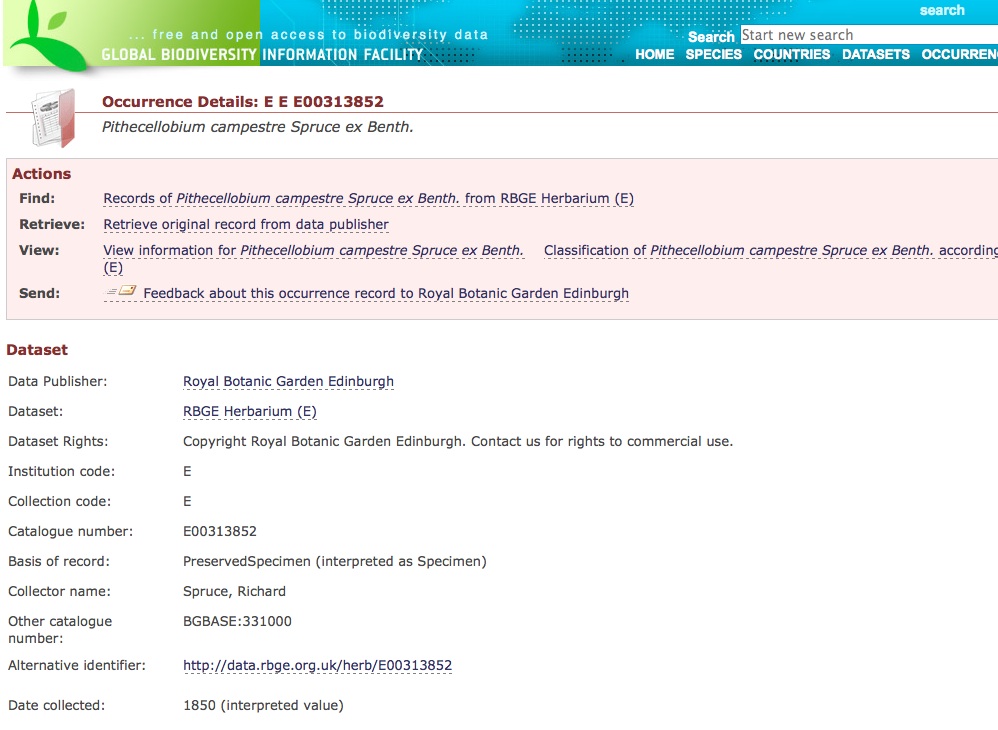
GBIF just gets simple records. Here is a screen shot of the page on GBIF for the specimen pictured above.
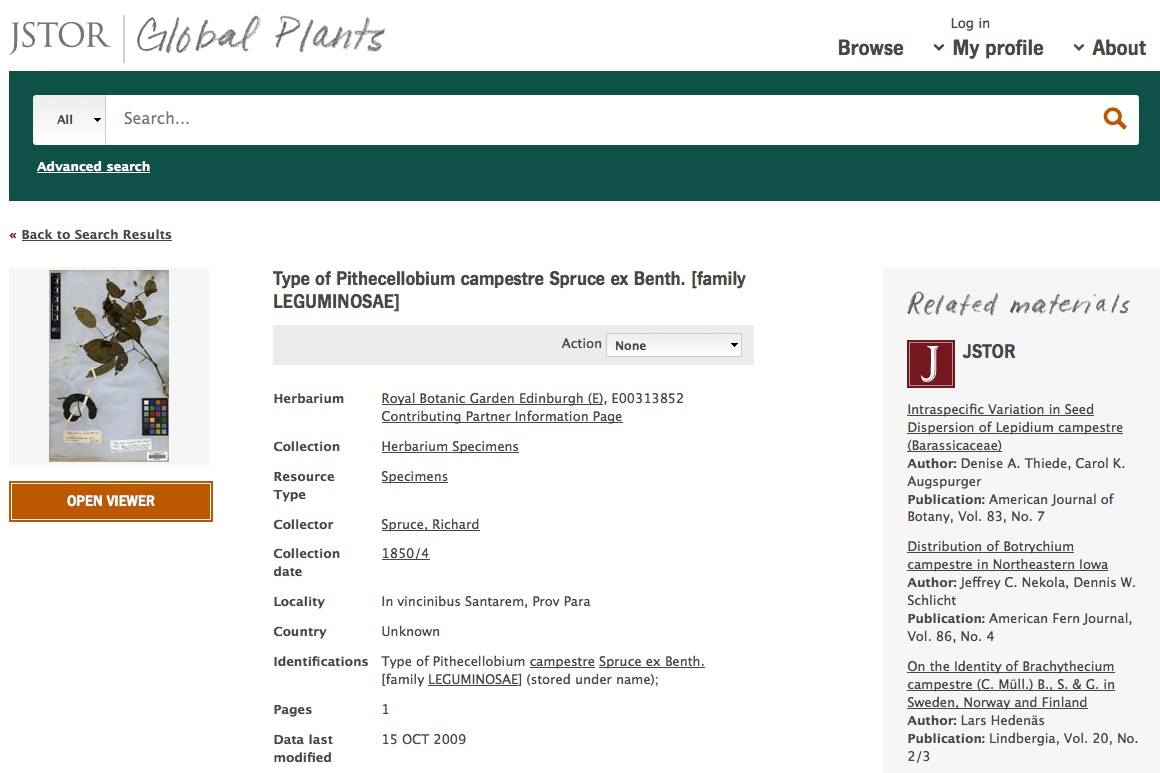
JSTOR gets images as well. Here is a screen shot of their page for the same specimen.
The same specimen may appear in other places like EoL (The Encyclopedia of Life) and we have our own herbarium catalogue as well of course.
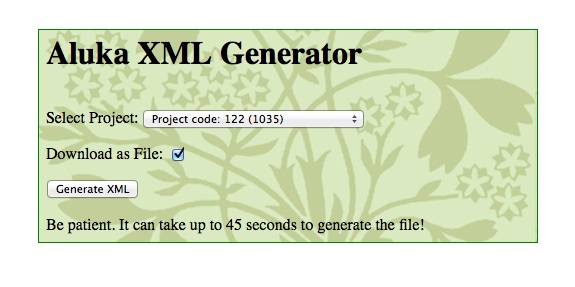
In order to keep the hundreds of thousands of records in all these different systems synchronized we need some programs to process the data. We couldn’t do it all by hand. This week I did some work on the program we use to export data to JSTOR. Batches of about 1,000 image files are bundled up with the data that accompanies them to be sent off. The data has to be in the right format. This program queries the databases and produces a file for a particular batch of images. I wrote it about a year ago and promptly forgot all about it but now it needed some changes. This is what the interface to the program looks like.
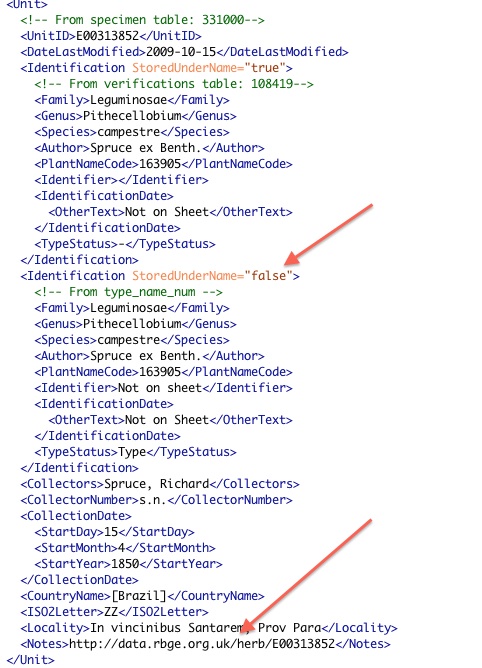
 OK – maybe it isn’t that exciting but at least it is prettier than the output! Here is a snippet of output – actually the bit about the specimen above.
OK – maybe it isn’t that exciting but at least it is prettier than the output! Here is a snippet of output – actually the bit about the specimen above.
I’ve added a couple of arrows to some of the parts that I changed this week. The lower one is the most important. We are starting to pass our specimen HTTP URIs to JSTOR so that they can link back to us so that their users have access to our original data. This was the subject of a workshop we held here at the botanics back in June. You might be able to see that GBIF are already including our specimen links in their pages.
If my fixes are OK then I will probably forget all about this particular program while Elspeth and Nicky use it to process thousands of specimens. This is just a small cog in a set of processes to enable biodiversity data to flow. There is a completely separate and far more complex system for handling the processing and storage of the images that Martin has developed. All these systems rest on other software components built by other people over many years.
This isn’t the only kind of thing I do at the office but perhaps it is one of the more important.