Life gets littered with untold stories; here’s one that did get told, briefly, and then got forgotten. It was told at the Botany meeting in Austin in August 2005. That summer Juan Carlos and I took a road trip to the Gulf coast with Argentinian friends Romina and Guillermo, driving along the beach of Padre Island, visiting the Alamo… to be honest, it’s hard to enjoy Texas in August when you’re over six months pregnant, feel like a whale, and the sticky heat never stops.
So, back to my Botany2005 talk, A molecular re-evaluation of diversity within Haplomitrium Nees (Marchantiophyta), with coauthors Barbara Crandall-Stotler and Ray Stotler (Southern Illinois University, Carbondale), David Long (RBGE) and Christine Davis, who was then at Duke University, North Carolina. The data set was never finalised, and the paper never written (and unlikely ever to be), so I’m going share some of the key points now instead.
We sampled roughly half of the accepted seven named species in the genus (Bartholmew-Began 1991), with a total of 11 accessions representing 4-6 species, and we used the liverworts Treubia and Apotreubia for the outgroup, based on the results of previous molecular phylogenetic analyses, and also on some shared morphological characters (tetrahedral apical cells, stalked slime papillae, and the calyptra or perigynium without perichaetial scales).
One thing that immediately struck us was the large amount of molecular sequence divergence within the genus Haplomitrium – in the following figure, we contrast it (in blue) with the molecular sequence divergence within the entire hornwort lineage (in red):
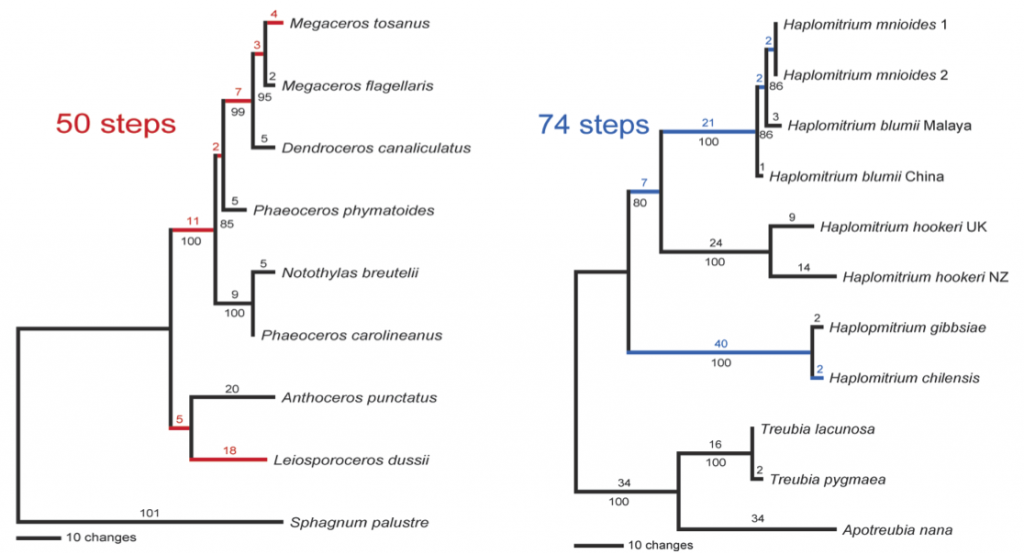
Phylograms produced from DNA sequence data from plastid internal transcribed spacer regions 2, 3 and 4, and the 4.5S and 5S rRNA genes; branch lengths are above the lines, parsimony bootstrap numbers are below. Hornworts on the left, Haplomitriopsida on the right.
The genus Haplomitrium contains more nucleotide diversity for the plastid internal transcribed spacer locus than the entire hornwort lineage does. In addition, Haplomitrium plastid internal transcribed spacer amplicon lengths are more variable – they range from 748 bases in the New Zealand Haplomitrium hookeri to 789 bases in Haplomitrium chilensis (range = 41 bases). Hornwort plastid internal transcribed spacer amplicon lengths range from 808 bases in Megaceros to 816 bases in Notothylas (range = 8 bases). (The commonly-used plastid trnL locus could not be used for this comparison, as it is largely unalignable across Haplomitrium.)
In order to generate a well-supported phylogeny for our accessions of Haplomitrium, we sequenced five plastid, one mitochondrial and two nuclear ribosomal markers.
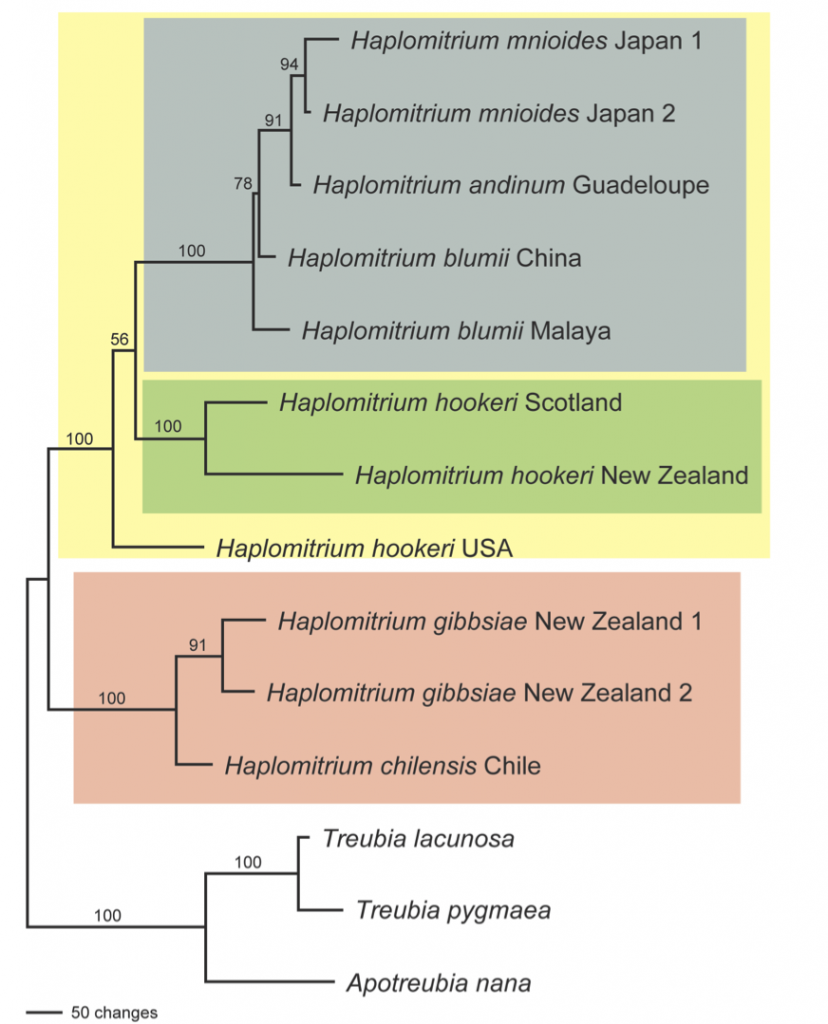
Phylogram for Haplomitrium based on plastid rbcL, rps4, trnL, atpB and psbA, mitochondrial nad5, and nuclear ribosomal SSU and LSU DNA sequence data; parsimony bootstrap values are shown above the branches.
Colours on the phylogram represent the sub-generic classification for Haplomitrium:
- Grey-blue = sect. Calobryum
- Green = sect. Haplomitrium
- Salmon = sect. Archibryum
The yellow box represents an unnamed lineage that forms a sister group to Archibryum, and includes one accession that does not fall into any of the sections. This is a plant from the Rockies of North America that had been identified as Haplomitrium hookeri (the only species known from the area) but that is clearly genetically very distinct from other plants that have been identified as such.
- Levels of morphological and DNA sequence divergence suggest that the separation of Haplomitrium into two genera would not be unwarranted
- Despite the small size of this group, and the fact that it has comparatively recently been comprehensively revised (Bartholmew-Began 1991), there are evidently still problems in species delimitation. High sequence divergence suggests that the UK and New Zealand H. hookeri could be recognised as separate; the US plant is even more clearly a distinct taxon.
In order to expand our sampling as far as possible, we looked at accessions of the genus that have been sequenced by other authors, and thus were publicly available to download from GenBank, and placed them on our phylogeny:
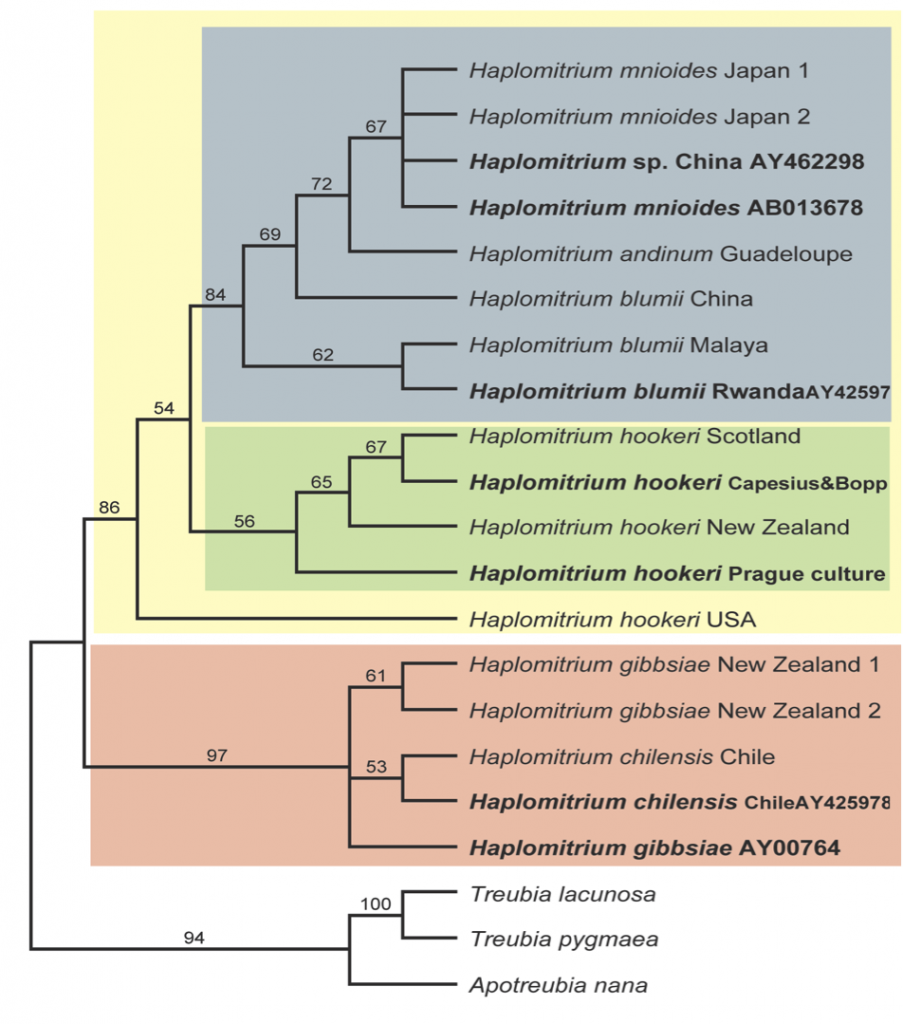
Phylogeny of Haplomitrium based on plastid rbcL, rps4, trnL, atpB and psbA, mitochondrial nad5, and nuclear ribosomal SSU and LSU DNA sequence data; parsimony boostrap values shown above the branches. Accessions in bold have been obtained from GenBank, and have only a subset of the DNA loci available, leading to reduced support for lineages.
However, when we started investigating the origin of some of these GenBank accessions, we discovered that even although the voucher information might superficially make it appear that plants were from different collections, seven GenBank accessions can be traced back to only two plants, both of which had been part of living culture collections. Sadly, due to high maintenance requirements, many of these collections are now lost.

Haplomitrium blumii (Nees) R. M. Schust. Axenic culture on agar, maintained in an environmental chamber at Southern Illinois University, Carbondale
Haplomitrium mnioides in GenBank: Out of five accessions on GenBank that were not associated with our own previous publications, the following….
- AJ09111 – Knoop, nad5 – “DNA from Y. Qiu”
- U87071 – Lewis et al. 1997, rbcL – “ML Sargent from Crandall-Stotler”
- AF231890 (rps4), AF231900 (trnL) – Newton et al. 2000 – “Crandall-Stotler culture”
….can all be traced back to a single accession, Furuki 7408 (Japan) (a plant that was in Barbara Crandall-Stotler’s culture collection at Southern Illinois University).
Haplomitrium hookeri in GenBank: Out of five accessions on GenBank that were not associated with our own previous publications, the following…
- AF313555 – Pryer et al. atpB – “ML Sargent no. H56”
- U87072 – Lewis et al. 1997; rbcL – “ML Sargent from WT Doyle, H56”
- U18504 – Hedderson et al. 18S – “Crandall-Stotler, living material”
…..are from a plant in Prague’s culture collection that was distributed in the US (but was lost from the Southern Illinois University culture collection) – we have not yet been able to identify where the Prague plant was collected, but it is not a 100% genetic match to the plants from Scotland, New Zealand and the US that we have sampled.
Acknowledgements: This study was largely financed by an NSF Partnerships for Enhancing Expertise in Taxonomy (PEET) award to Barbara Crandall-Stotler (DEB-9977961). Plant material was kindly shared by Drs Bernard Goffinet (University of Connecticut), Tatsuwo Furuki (Natural History Museum and Institute, Chiba), Jeff Duckett (Natural History Museum, London), John Engel and Matt von Konrat (Field Museum, Chicago), Wilfred Schofield (UBC), and K. Katoh.

