Sphaerocarpos texanus and S. michelii, photographed by David Long and Fred Rumsey for the British Bryological Society Field Guide (see references for link)
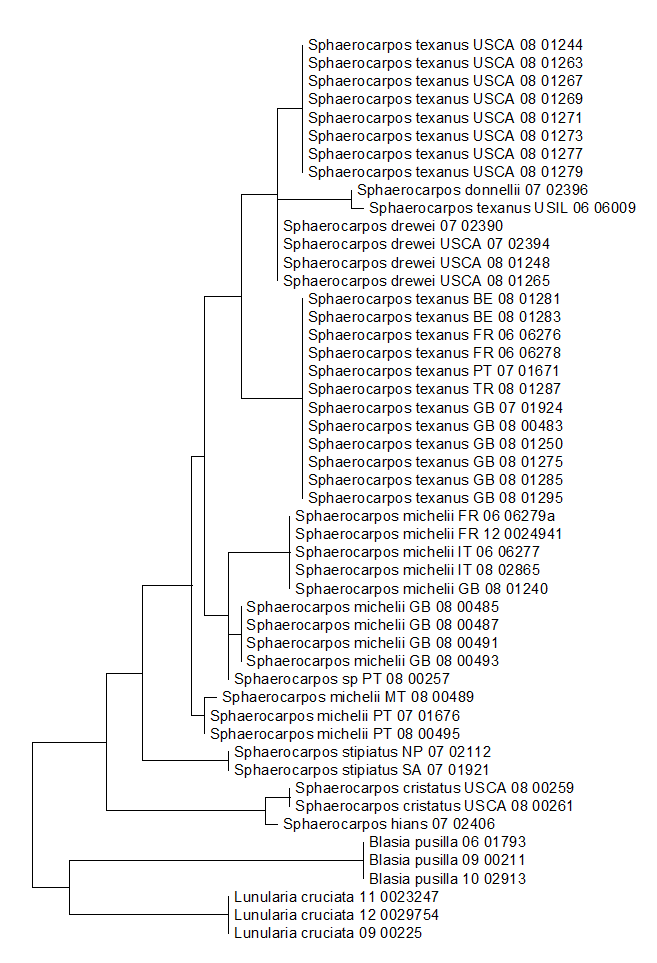
In conjunction with Dr Daniela Schill’s monographic work on Sphaerocarpos, we’ve been building a molecular phylogeny for the genus. We have attempted to extract DNA from 66 accessions, including three S. cristatus, all from California, seven S. donnellii from the US, five S. drewei from California, two S. hians, 13 S. michelii from France, Great Britain, Italy, Malta and Portugal, two S. muccilloi, five S. stipitatus from Nepal, Portugal and South Africa, and 25 S. texanus from Belguim, France, Great Britain, Italy, Portugal, Turkey, California and Illinois. We have also included some as yet unidentified material, including an accession from Chile.
Because much of our work at RBGE focuses on plant DNA barcoding and the protocols are established and frequently successful, we have chosen to use sequence data from some of these barcoding regions for this project. However, the liverwort matK primer sets were not very successful in Sphaerocarpos, with a very limited number of good quality sequences generated. The nuclear ITS2 region had its own issues, with many of the sequence reads being difficult to interpret due to overlapping peaks. In the end we focused on the three most successful plastid loci, the rbcL and rpoC1 barcoding amplicons, and a region that encompasses part of the psbA gene and the psbA-trnH intergenic spacer.
Unfortunately, we have not been able to amplify DNA from all the samples we extracted, with failures particularly for some of the older specimens. One of the species we attempted to sequence, Sphaerocarpos muccilloi, has not worked for any of the gene regions that we have been using, while another species, Sphaerocarpos hians, has so far only amplified for a single region, rbcL.
Although each species seems to be genetically distinguishable from the other species sampled, two of the most widespread species, S. michelii and S. texanus, are resolving as para- or polyphyletic. The phylogenetic tree contains three distinct groups of S. michelii accessions, and two distinct groups of S. texanus, one from Europe and the other from California. An Illinois accession that has been published as S. texanus resolves here with S. donnellii. The Illinois material lacks spores and is thus difficult to identify morphologically, but is outwith the Southeastern Coastal Plain area where S. donnellii is thought to occur.
The next steps in this study involve a second pass through the DNA extractions, to see if using other PCR additives will help increase the sequence success rate, then combining the sequence data from the three sequenced loci into a single matrix, to produce a more robust and supported phylogeny. Description of new species, where required, will fall under Daniela’s remit, in line with the comprehensive taxonomic revision that she has carried out.
Links & References:
Sphaerocarpos, preview to a monograph