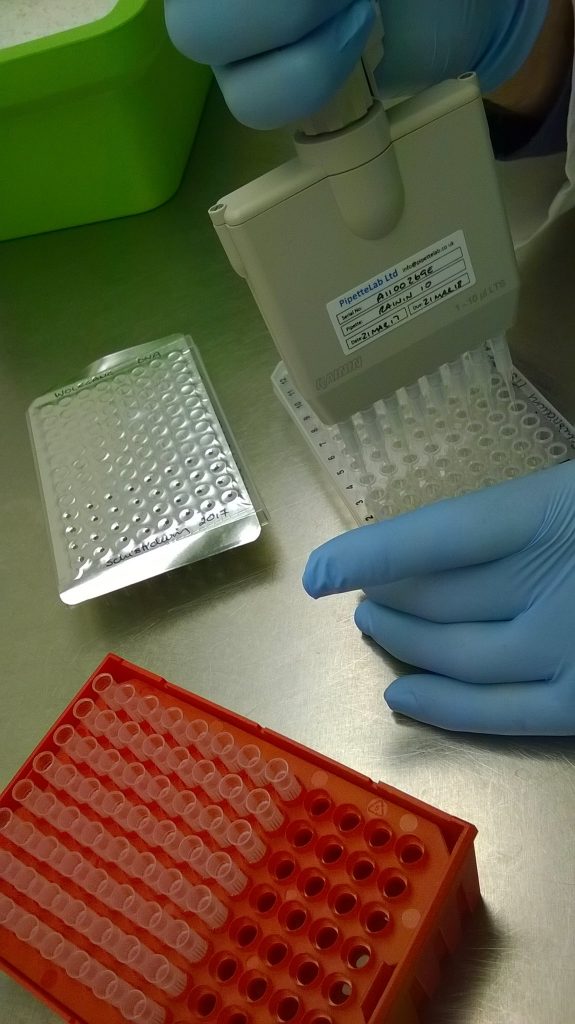
After we extracted a plate’s worth (12 columns by 8 rows, or 96 samples) of Schistidium DNA, the next step in our process is to copy a preselected part of that DNA, using the Polymerase Chain Reaction (PCR). For this study with Wolfgang, we are copying a region of nuclear DNA known as the Internal Transcribed Spacer 2 (ITS2). We set up the reactions in a laminar flow hood, which blows clean air down onto the work surface, keeping everything as clean as possible. Most of the reagents are kept in a -20ºC freezer between uses, so these get set out to defrost before use (time for a coffee break!). The DNA polymerase enzyme, taq, on the other hand, is stored in glycerol so doesn’t freeze at -20ºC; while we’re using it, we keep it on ice so it stays cold.
 The reaction components are added into a single “Master-Mix” tube, which includes water, buffer, Magnesium, enhancing additives, short oligonucleotides (primers), and the taq enzyme. Small (19 μl) aliquots of this master mix are then added to a new 96-well plate, and 1 μl of DNA from the extraction plate is transferred across into each of the 96 reactions.
The reaction components are added into a single “Master-Mix” tube, which includes water, buffer, Magnesium, enhancing additives, short oligonucleotides (primers), and the taq enzyme. Small (19 μl) aliquots of this master mix are then added to a new 96-well plate, and 1 μl of DNA from the extraction plate is transferred across into each of the 96 reactions.
Once the DNA has been added, the plate is sealed (this time with a clear plastic film that sticks firmly in place when heated), briefly spun in a centrifuge to make sure all the reagents are mixed together at the bottom of the 96 plastic wells, and transferred into one of our Thermocyclers, or PCR machines.
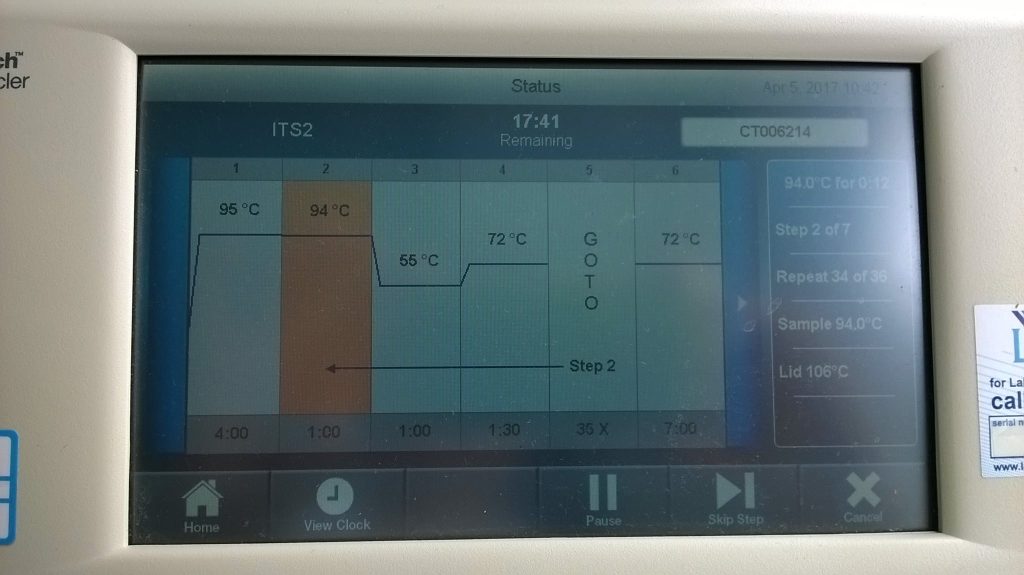
The plate sits in a metal block which rapidly “cycles” up and down in temperature, following a predetermined programme. For the gene region that we are copying here, the programme first heats the block to 95ºC for 4 minutes, then starts a cycle of 94ºC for 1 minute, 55ºC for 1 minute, and 72ºC for 45 seconds, repeated 30 times. When the block is at 94ºC, the double-stranded DNA is pulled apart into single strands; when it’s cooled to 55ºC the primers stick on and the taq initiates making copies of the ITS2 region, and when it is heated up again to 72ºC, the copies get completed, before they’re pulled apart again when the block heats up to 94ºC and the cycle starts again…
A couple of hours later, the reaction is complete, and at this point we HOPE that all the 96 wells in our plate contain millions of copies of the ITS2 DNA molecule from the Schistidium extraction that was added in each case. However, in order to see if any of it has actually worked, we need to stain and visualise the DNA, and for this, we have to run a gel, a process that will be the subject of the next installment.
Links to reports on Moss diversity in an artificial landscape, an EU Synthesys Access project with Dr Wolfgang Hofbauer at RBGE:
- Building on building mosses, a return to Schistidium in the built environment http://stories.rbge.org.uk/archives/24310
- Volunteering at the Botanics – bryophytes in our living landscape http://stories.rbge.org.uk/archives/24333
- Campylopus introflexus, an invasive alien on the glasshouse roof http://stor/ies.rbge.org.ukarchives/24359
- The trials and tribulations of a moss in the lab: DNA extraction http://stories.rbge.org.uk/archives/24399