When working out new protocols, it’s very useful to have a big stash of DNA to test them on. Just now, in a collaborative project with Dr Michelle Hart (Scientific and Technical Services), Drs Mark Hughes and Catherine Kidner (Tropical section), and RBGE PhD student Hannah Wilson, we’re investigating some of the ways that the quality of DNA extracted from herbarium specimens relates to how the plant was treated in the field.
While Hannah has been extracting DNA from some of the wild New Guinean plants that she’ll be working with for her PhD, I’ve been working with some material from rather closer to home. From Mark’s office, two doors along the corridor, in fact…
Just before lunch on Thursday, a house-plant that’s been surviving on an office shelf donated a couple of its young leaves to science. I popped them into a mortar with a dash of acid-washed sand and some extraction buffer, and ground them up, breaking open the plant cell walls so that the DNA could be washed out into solution.
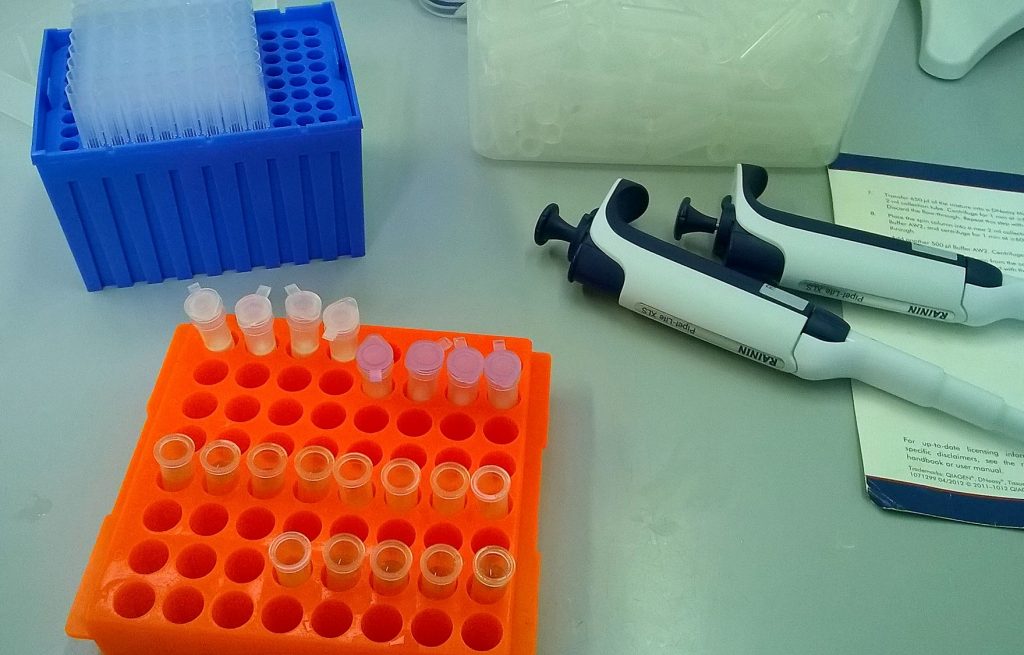
QIAGEN plant DNeasy mini-column extractions in progress; I put everything through the same four QIAshredder spin columns (purple), but ended up eluting into six DNeasy mini spin columns (white).
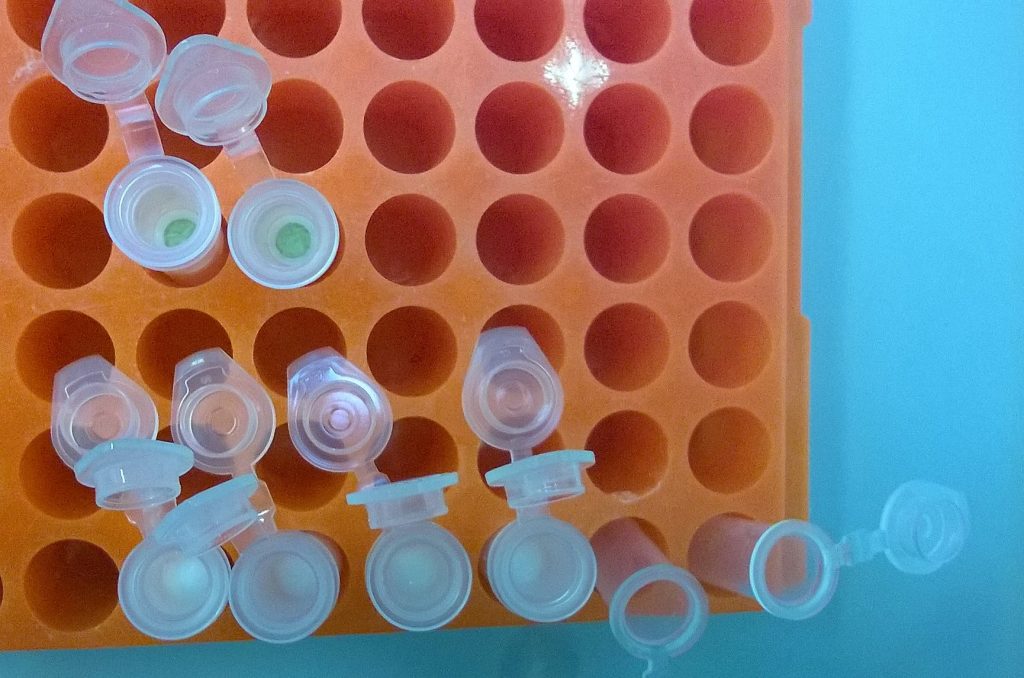
QIAGEN plant DNeasy mini spin columns with the Begonia DNA bound to the column membranes; the upper two columns have not yet been washed, and the membrane has been stained green by the Begonia extraction; an ethanol wash step has cleaned the membrane of the loser four columns.
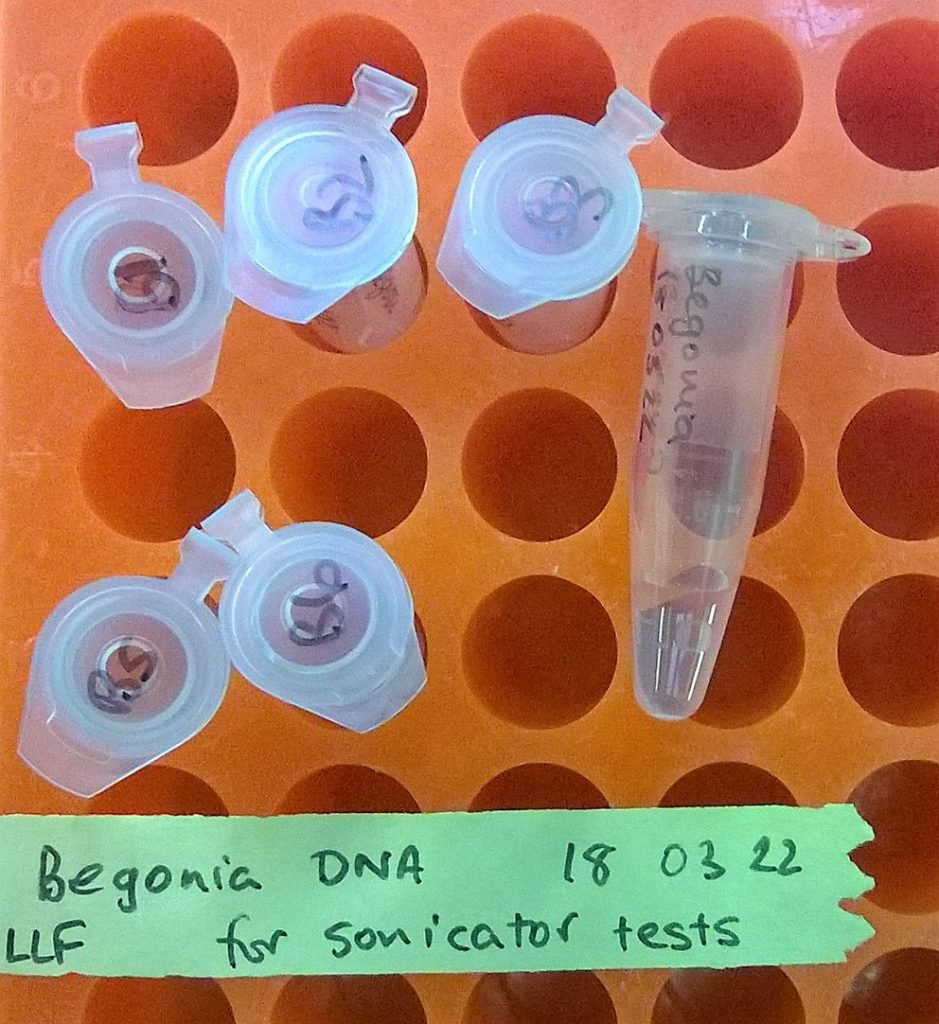
Six microcentrifuge tubes that (hopefully) now contain high molecular weight Begonia DNA extractions…
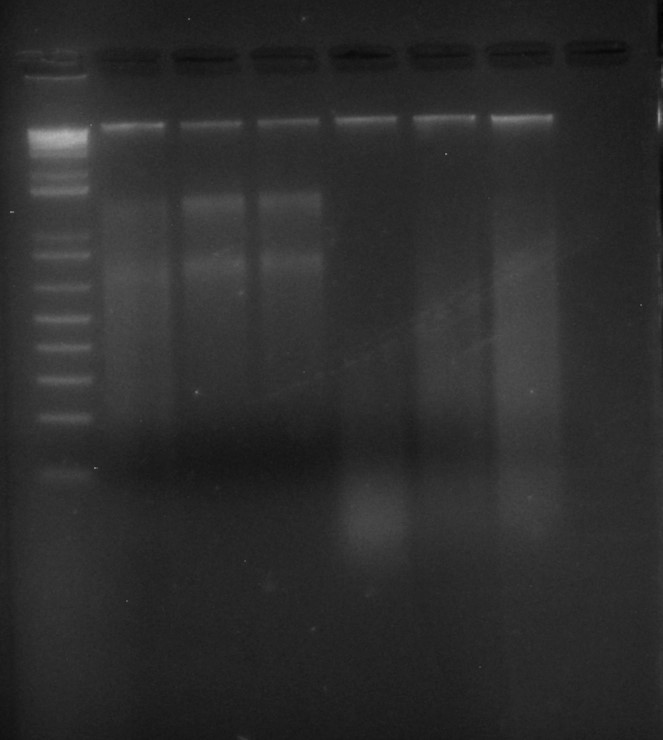
And the evidence that it all worked: LANE 1: 3.5 ul of Invitrogen 1kb ladder; LANES 2-7: 6 ul of Begonia DNA extractions, run on a 2% agarose TBE gel for 60 mins at 80 volts.
All six tubes contain high molecular weight DNA (the nice sharp bright band near the top of the picture); rather oddly, the first three also contain clear bands of RNA, while the second three, which were treated identically, do not.
As we’re planning to use this DNA over the next couple of weeks, I’ve put the tubes in our lab fridge rather than popping them into the freezer, to avoid a freeze-thaw cycle that might damage the DNA. I’ll need to get a rather more accurate picture of the DNA quality and quantity before we start testing protocols with it, so will run a Tapestation genomic DNA tape, and also get readings from our deNovix. I’ll leave off doing this until just before we use the DNA, though, particularly as it’s not being stored in lo-bind tubes.



