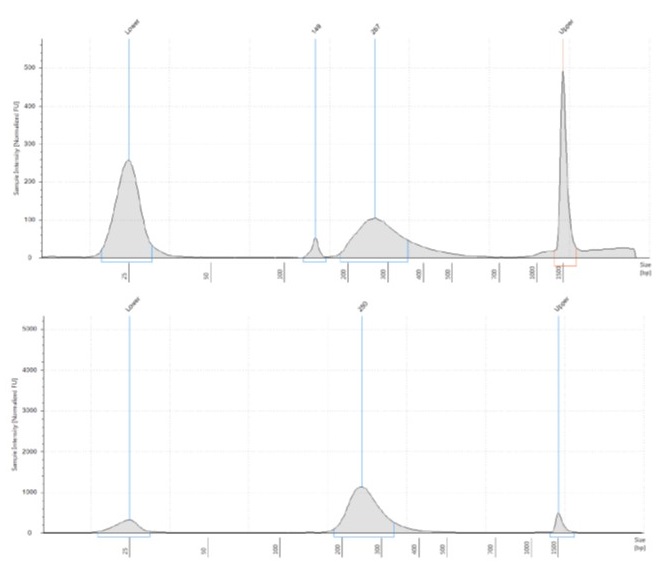
After testing bead-to-sample ratios of 30:50, 35:50, 40:50, 50:50 and 60:50 on a Thermo Scientific™ GeneRuler™ 50 bp DNA Ladder, using Beckman Coulter AMPure XP beads, we focused on the 50:50 (or 1:1) ratio, deciding to test it on a real pool of hybrid-captured libraries that we were not intending to use for sequencing (having rather overdone the post-capture amplification). There was enough DNA in the pool to split it into three 50 µl aliquots, allowing us to try 40:50, 50:50 and 55:50 bead-to-sample ratios.
We ran the cleaned samples on an Agilent High Sensitivity D1000 ScreenTape.
All three of the bead-to-sample ratios tested removed most of the adaptor peak.
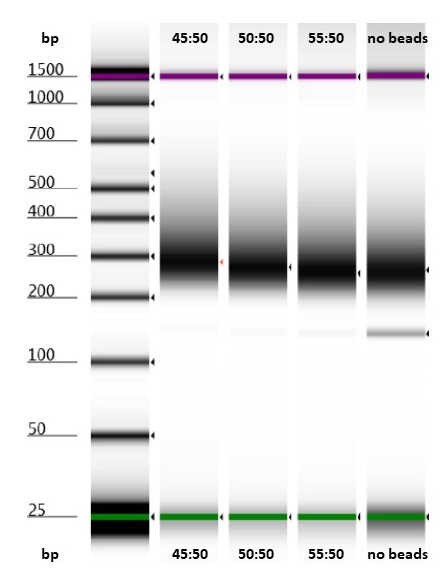
Hybridisation Pool 4, with 20 cycles of post-capture PCR: bead-to-sample test for removing adaptor peaks. The adaptor peak is clearly visible in the “no beads” lane, and faintly visible in the other lanes.
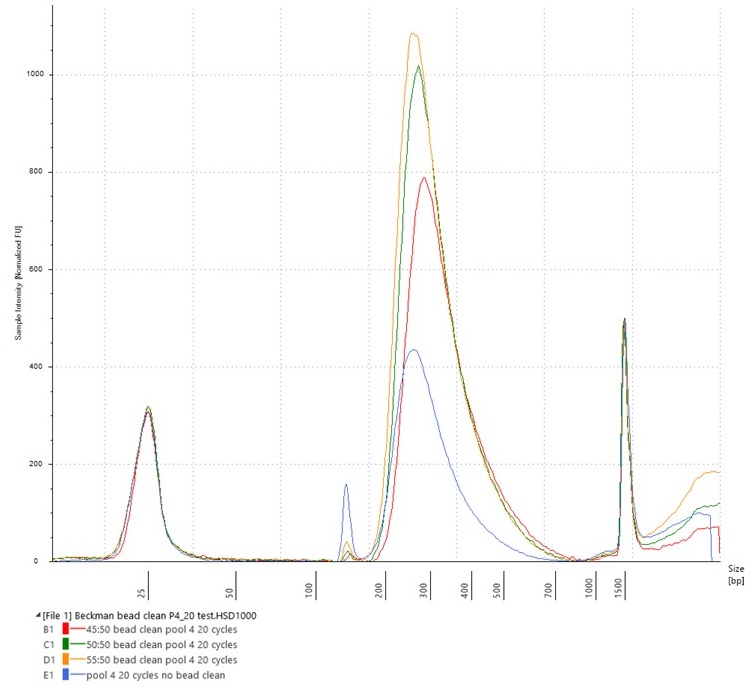
Hybridisation Pool 4, with 20 cycles of PCR (blue), with overlay of the test of different bead-to-sample ratios
The bead-cleaned library pools (visible as red, green and yellow lines) have far higher peaks (i.e., more concentrated DNA) at the c. 300 bp library size than the original Pool 4 (visible as a blue line), having been eluted into a smaller volume. They all also have far lower adaptor peaks.
In the end we used a 1:1 ratio of bead-to-sample to reduce the adaptor peaks in the libraries that we used for sequencing.
Hybrid capture from degraded DNA: testing bead cleaning on a 50bp ladder