When we made our Begonia libraries, working with (in some cases) relatively small quantities of very degraded DNA, we should have diluted the adaptors. We didn’t. Consequently, we ended up with huge adaptor peaks in some of our libraries. Meanwhile the clock was ticking and we really needed to get the lab-work finished and the samples sent off for sequencing.
The obvious thing to do was a bead-clean to try to remove as much of the c. 150 base adaptor peaks, while leaving as much of the actual libraries, as possible. So, what bead-to-sample ratio would be best….?
Following a tip from Dr Linda Neaves, we used some Thermo Scientific™ GeneRuler™ 50 bp DNA Ladder as a proxy for the “library sample” to be bead-cleaned, and tested five different bead-to-sample ratios on it:
A 5 ml aliquot of beads was removed from the fridge about half an hour beforehand, to come to room temperature, and thoroughly vortexed before use to resuspend the beads. The beads were mixed with the sample (i.e. the ladder) in 1.5 ml eppendorf tubes, by pipetting slowly up and down a few times; the mixture was left for 5 minutes at room temperature, then 5-8 minutes on a magnetic rack for the beads to form a pellet; the supernatant was discarded. The beads were washed twice with 200 µl of 80% ethanol, then air dried for up to 5 minutes, removed from the rack and resuspended in 30 µl 0.1x TE to elute the DNA into the buffer. The samples were left for two minutes, then placed back on the magnetic rack for 2-5 minutes. The supernatant was removed, and 2 µl of each sample was run on a Tapestation on an Agilent High Sensitivity D1000 ScreenTape.
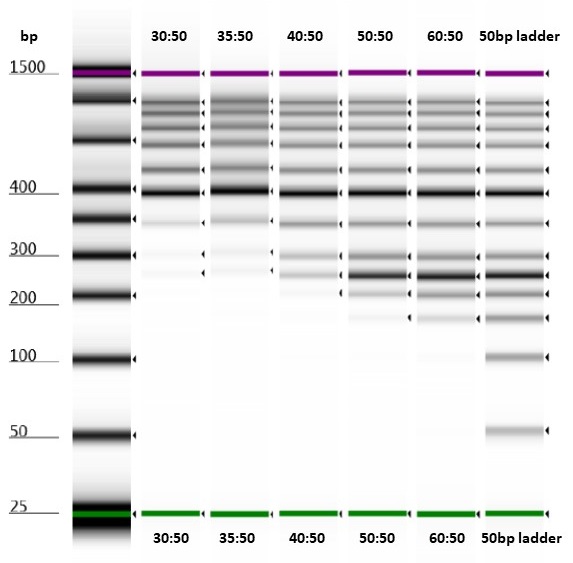
Test of varying bead-to-sample ratios on a Thermo Scientific™ GeneRuler™ 50 bp DNA Ladder run on an Agilent High Sensitivity D1000 ScreenTape.
Another way of viewing this is by overlaying all the bead-cleaned sample traces on the uncleaned ladder (visible as the light green peaks below):
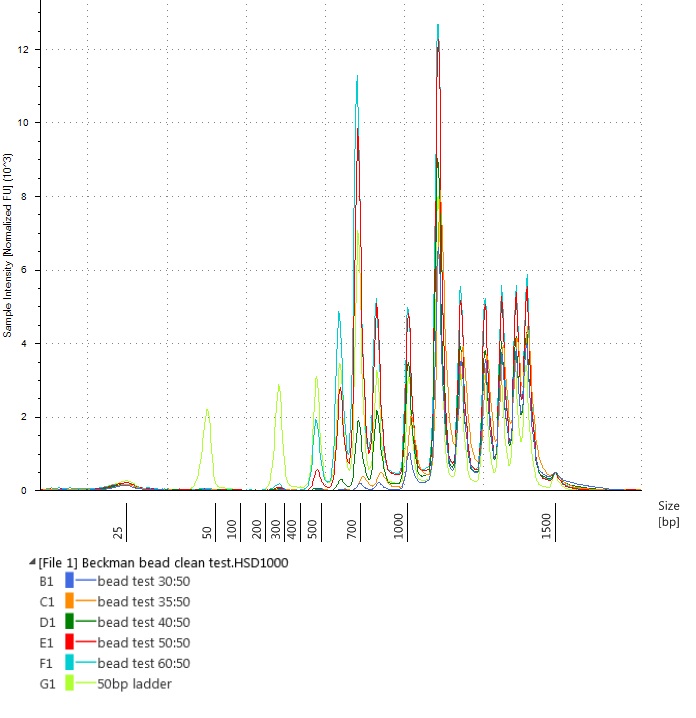
Test of varying bead-to-sample ratios on a Thermo Scientific™ GeneRuler™ 50 bp DNA Ladder – overlaid traces from the Agilent High Sensitivity D1000 ScreenTape run
All the bead-to-sample ratios that we tried successfully removed short fragments (around the size of the adaptor peaks) from our samples. However, it seemed that approximately equal volumes of bead-to-sample would remove less of the library-sized DNA fragments.
As such, it was a useful test, and allowed us to go on to test the bead cleans on a surplus pool of hybridised libraries, just to make sure that we had the ratios right, before cleaning the adaptors out of our sequencing pools.
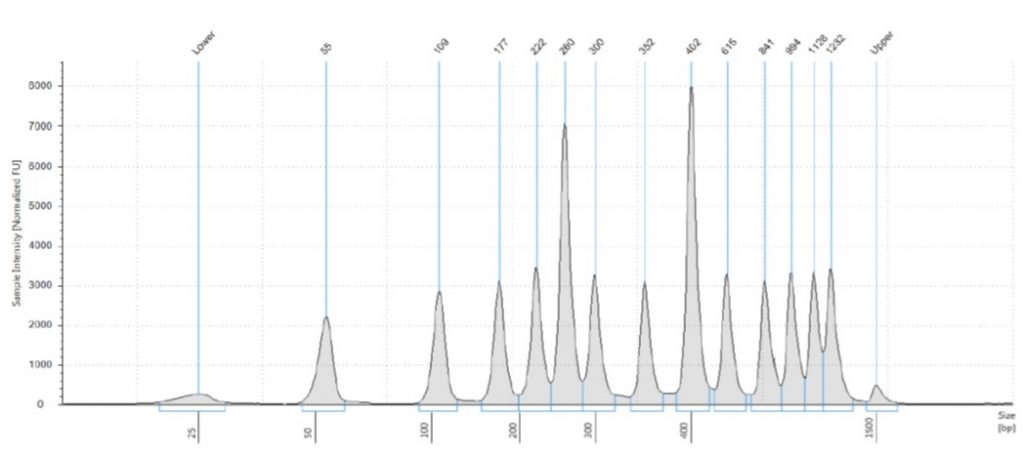
The less welcome news, however, was that the Tapestation software did rather an inconsistent job of assigning sizes to the bands in the 50 bp ladder. The Tapestation sizing of the bands was:
55 bp (for the 50 bp band),
109 bp (for the 100 bp band),
177 bp (for the 150 bp band),
222 bp (for the 200 bp band),
260 bp (for the 250 bp band),
300 bp (= the 300 bp band),
352 bp (for the 400 bp band),
402 bp (for the 500 bp band),
615 bp (for the 600 bp band),
841 bp (for the 700 bp band),
994 bp (for the 800 bp band),
1126 bp (for the 900 bp band),
1232 bp (for the 1000 bp band).


Chiara Barozzi
Dear,
I found your post very interesting and useful, I would like to repet this trial with some new clean beads other than Ampure, so I would like to know the amount of ladder DNa you used.
thanks in advance,
best regards
Chiara Barozzi
Laura
Oh dear – I’m afraid not only did I not include the ladder concentration in the story, but I can’t think where I’d have written it down… what an annoying omission! I’ll let you know if I find it anywhere.