The mantra for many years for next generation sequencing has been, like “garbage in, garbage out”, that the optimal starting point is high quality, high molecular weight DNA. For many of my favourite plants, getting this DNA, and not the actual sequencing, is the hardest part of the project. It’s a bit upsetting, then, when the first step in next generation library preparation, for the methods that we have been using, is to fragment that hard-won DNA into relatively small pieces, and then throw away any of it that ends up too big or two small.
Particularly when dealing with herbarium samples, obtaining sufficient DNA can be problematic. Luckily the pipeline for hybrid capture – breaking the DNA into suitable fragment sizes, sticking on some adaptor DNA, fishing out the interesting fragments from the total genomic DNA “soup”, and copying those interesting fragments a few times before sequencing them – does allow for relatively small starting quantities of DNA. We tested two different kits in our NBAF project. The first is the Illumina Tru-Seq Nano library preparation kit (FC-121-4001), which recommends a starting DNA quantity of 100 ng. We also used NEB‘s NEBNext Ultra library preparation kit for Illumina (E7370S), which has been optimized for as little as 5 ng of starting DNA.
There are a variety of fragmentation methods: for 454 libraries I have used nebulization, which effectively involves passing the DNA through a small pore; Illumina‘s Nextera kits use enzymatic degradation (a method that seems to be unsuited to MYbaits hybrid capture for plant DNA, possibly due to non-random action of the enzymes – pers. commun. James Nicholls); for TruSeq, Illumina recommend fragmentation by sonication. Nicholls et al. sonicated their silica-extracted Inga DNA using Diagenode’s Bioruptor Plus, a protocol that we have followed (where required…)
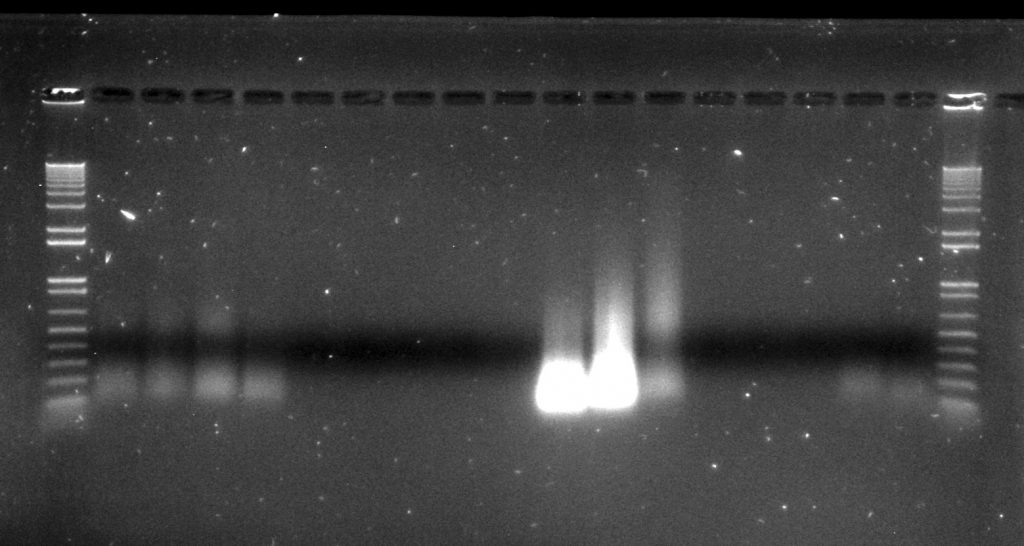
Inga umbellifera DNA extractions from FDGB 1948 (4 lanes), Hostmann 1840 (5 lanes), Lawrance 1932 (3 lanes), Tello 1991 (3 lanes), and Matthews 1835 (2 lanes), surrounded by Invitrogen’s 1kb ladder
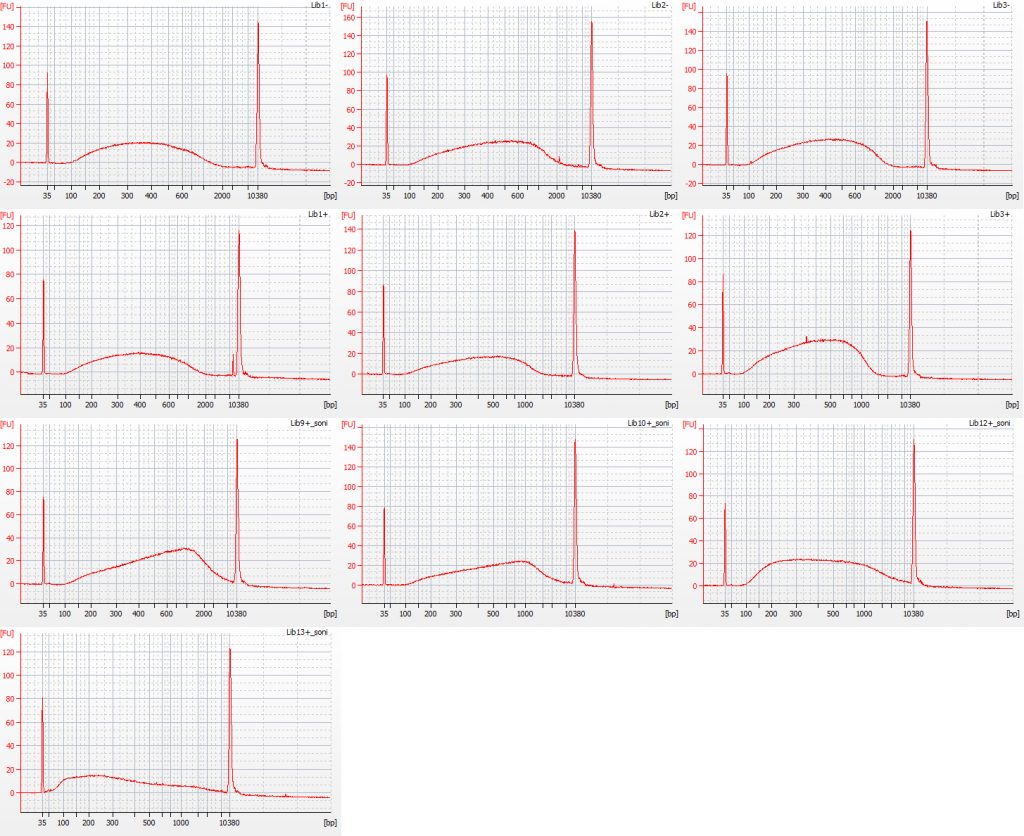
However, after being collected, and dried out, and possibly heated, and shipped from the tropics back to Edinburgh, and stuck on a sheet of paper, and possibly frozen and fumigated and goodness knows what else over the years they have spent in the herbarium, none of the herbarium samples that were more than 11 years old that we included in our study actually contained any measurable high molecular weight DNA (the 1932 leaf material was actually best preserved, with some fragment sizes of over 3,000 bp, as estimated by our Tapestation). In essence, the passage of time has performed our fragmentations for us.
Because some of the herbarium material did contain DNA fragments that were longer than the 350 bp fragment size that we were aiming for, we performed some sonication on some samples. This was tailored to the fragment size profile of each DNA sample, with the highest molecular weight DNA sheared, following the Tru-Seq 350 bp insert size protocol, on low power for 8 cycles of (30 seconds on, 90 seconds off), in 1.5 ml tubes (Nicholls et al. submitted).
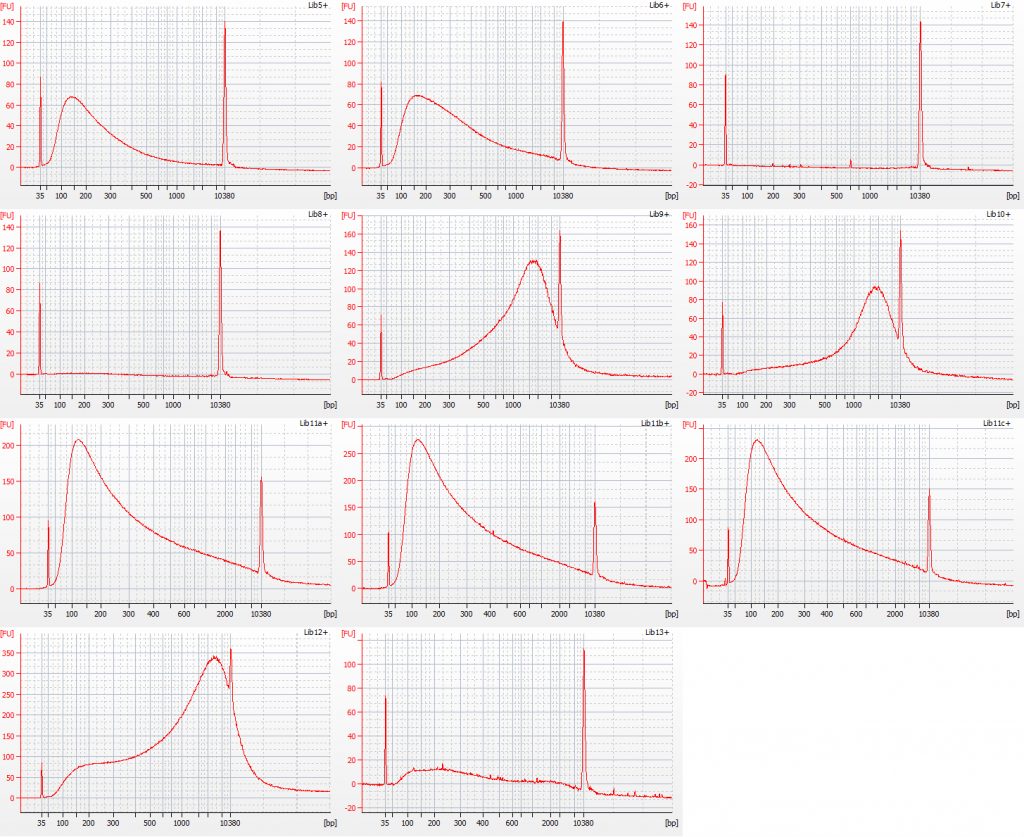
Normal sonication (8 cycles) was used for the unrepaired and repaired 2009 silica and herbarium DNAs.
Modified sonication was used for the 2004 herbarium DNA and the 1932 herbarium leaf DNA (3 cycles) and the 1835 herbarium DNA (1 cycle).
No sonication was used on the 1948 herbarium DNA (1st and 2nd plots, below), the 1932 herbarium flower DNA (three aliquots – 7th-9th plots, below), and the 1840 herbarium DNA (3rd and 4th plots, below).
Bioanalyser traces for unfragmented repaired Inga umbellifera DNA from 1948, 1948, 1840, 1840, 2004, 2004, 1923 flower a, 1932 flower b, 1932 flower c, 1932 leaf, 1835
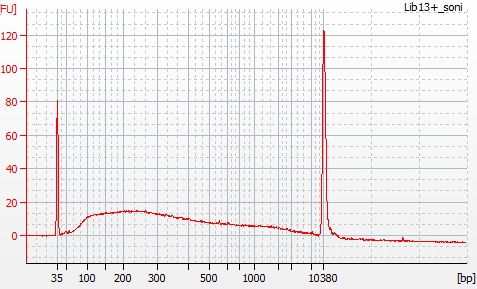
Above: pre-sonication Inga umbellifera DNA fragments. Below: post-sonication Inga umbellifera DNA fragments.
Bioanalyser traces for Inga umbellifera (top left to bottom right): 2009 herbarium material, unrepaired, 8 cycles sonication; 2009 silica material, unrepaired, 8 cycles sonication; 2009 silica material, unrepaired, 8 cycles sonication; 2009 herbarium material, repaired, 8 cycles sonication; 2009 herbarium material, repaired, 8 cycles sonication; 2004 herbarium material, repaired, 3 cycles sonication; 2004 herbarium material, repaired, 3 cycles sonication; 1932 herbarium leaf material, repaired, 3 cycles sonication; 1835 herbarium material, repaired, 1 cycle sonication
Following the shearing of high molecular weight DNAs using sonication, the Illumina Tru-Seq Nano protocol includes a bead clean-up step. However, most bead- or column-based clean-ups involve at least a degree of sample loss, and James Nicholls has already calculated that about 30% of the original DNA is lost at this stage. Therefore, for the recommended 100 ng of DNA that is sonicated for an Illumina Tru-Seq Nano library, only around 70 ng remains after the bead clean-up step. We skipped the bead clean-up completely for one of our sonicated DNAs (1835 herbarium material) because we had so little DNA.
James A. Nicholls, R. Toby Pennington, Erik J.M. Koenen, Colin E. Hughes, Jack Hearn, Lynsey Bunnefeld, Kyle G. Dexter, Graham N. Stone & Catherine A. Kidner. 2015. Using targeted enrichment of nuclear genes to increase phylogenetic resolution in the neotropical rain forest genus Inga (Leguminosae: Mimosoideae). Frontiers in Plant Science 6: 710. doi: 10.3389/fpls.2015.00710
Capturing Genes from Herbaria. I. What it’s all about. http://stories.rbge.org.uk/archives/16411
Capturing Genes from Herbaria. II. Inga. http://stories.rbge.org.uk/archives/16427
Capturing Genes from Herbaria. III. The samples. http://stories.rbge.org.uk/archives/16441
Capturing Genes from Herbaria. IV. DNA. http://stories.rbge.org.uk/archives/16470
Capturing Genes from Herbaria. V. Fragmenting the DNA. http://stories.rbge.org.uk/archives/16525
Capturing Genes from Herbaria. VI. Size Selection. http://stories.rbge.org.uk/archives/16645
Capturing Genes from Herbaria. VII. Comparisons. http://stories.rbge.org.uk/archives/16737
Capturing Genes from Herbaria. VIII. Amplification. http://stories.rbge.org.uk/archives/16788
Capturing Genes from Herbaria. IX. Hybrid capture. http://stories.rbge.org.uk/archives/17298
Capturing Genes from Herbaria. X. An update. http://stories.rbge.org.uk/archives/20751
Capturing Genes from Herbaria. XI. Some metagenomics of a herbarium specimen. http://stories.rbge.org.uk/archives/20817