In March this year, having already chosen and obtained the plant material that we were going to use for our NBAF project on using a hybrid bait protocol to recover DNA sequences from herbarium material, Michelle Hollingsworth and I started on the DNA extractions.
Although there were only seven accessions of Inga umbellifera, multiple DNA extractions were made from each accession, to have enough material to make replicate libraries, and to be able to treat some of the DNA in different ways. In addition, we did not expect to get very high yields of DNA out of some of the older herbarium samples, so made multiple extractions in case we had to pool the DNA downstream.
The plant tissue was ground up in 2ml tubes, using a TissueLyser II with flattened tungsten beads. Afterwards, most DNA was extracted using a standard protocol for Qiagen DNeasy Plant mini-kits, with the exception that several sets of extractions (lysate plus buffer AW1) were combined through single white DNeasy Mini spin columns, in order to concentrate the DNA. A second set of 5 samples were extracted following standard protocols for the Qiagen DNeasy Plant mini-kit, but instead capturing and eluting the DNA from Qiagen QiaQuick PCR purification columns, which have been suggested to perform better at eluting small DNA fragments.
2009: no. 16L 145, FRENCH GUIANA (E):
3 Plant DNeasy extractions from silica-dried material
1 Plant DNeasy + QiaQuick extraction from silica-dried material
3 Plant DNeasy extractions from herbarium material
1 Plant DNeasy + QiaQuick extraction from herbarium material
2004: Kyle Dexter no. 401, PERU (E)
3 Plant DNeasy extractions from herbarium material
1 Plant DNeasy + QiaQuick extraction from herbarium material
1991: Tello 2608, PERU (E)
3 Qiagen Plant DNeasy extractions from herbarium material
1948: FDBG no. 5682, GUIANA (K)
7 Plant DNeasy extractions from herbarium material
1 Plant DNeasy + QiaQuick extraction from herbarium material
1932: Lawrance no. 260, COLOMBIA (E)
3 Plant DNeasy extractions from herbarium material: 1 from flower fragments, 1 from random plant parts, and 1 from leaf fragments
1840: Hostmann no. 1711, SURINAME (K)
7 Plant DNeasy extractions from herbarium material
1 Plant DNeasy + QiaQuick extraction from herbarium material
1835: Matthews no. 1593, PERU (E)
2 Plant DNeasy extractions from herbarium material
5μl of each DNA extraction was mixed with a bromophenol blue loading dye and run on a 1% agarose TBE gel, stained with SybrSafe. DNA was also quantified using Qubit dsDNA high sensitivity kits.
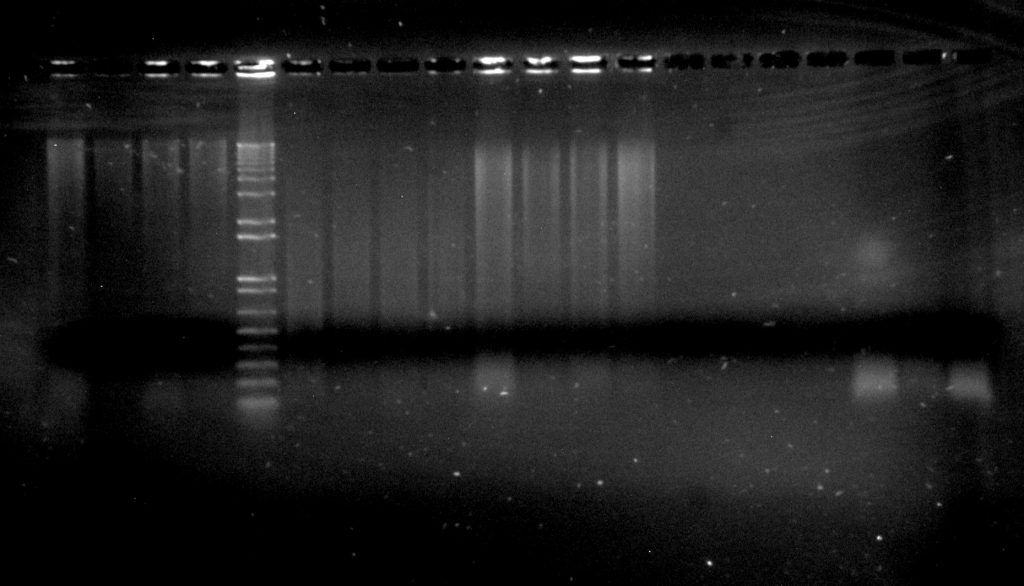
Inga umbellifera DNA extractions: 16L-145 2009 silica (4 lanes); Invitrogen 1kb ladder; Dexter 2004 herbarium (4 lanes); 16L-145 2009 herbarium (4 lanes); Hostmann 1840 (4 lanes); FDGB 1948 (3 lanes)
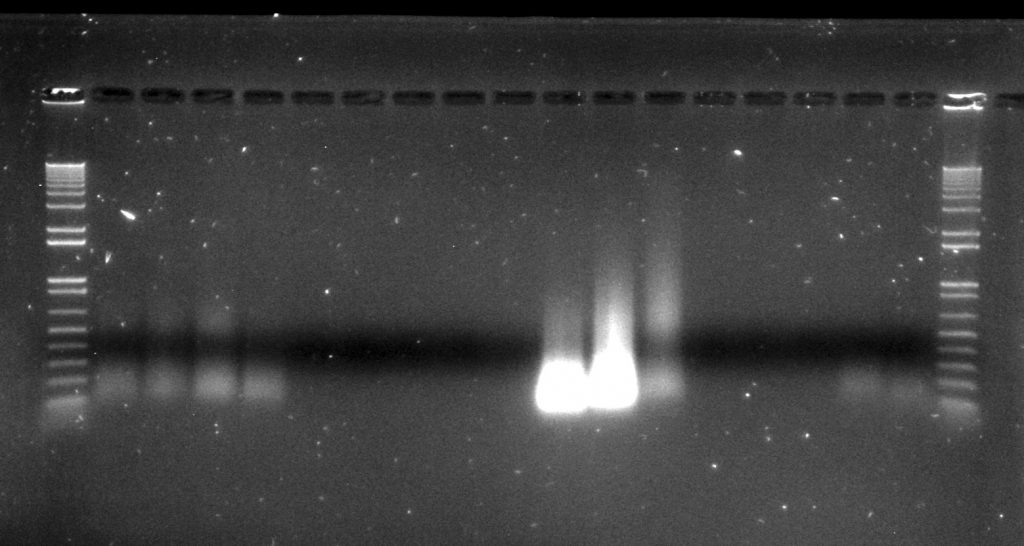
Inga umbellifera DNA extractions from FDGB 1948 (4 lanes), Hostmann 1840 (5 lanes), Lawrance 1932 (3 lanes), Tello 1991 (3 lanes), and Matthews 1835 (2 lanes), surrounded by Invitrogen’s 1kb ladder
A single specimen, Tello 2608, gave such low DNA yields (of the three extractions, two were too low to measure with the Qubit dsDNA high sensitivity kit, while the third measured 0.053 ng/μl) that it was excluded from library preparation. Surprisingly, at 24 years old, this is one of the most recent herbarium specimens in our study.
Because degraded DNA can have small breaks in the strands, but is double stranded when extracted, subsequently denaturing the extracted DNA in order to perform PCR can result in it fragmenting into even smaller pieces, as the strands separate. We repaired most of our DNA extractions using NEB’s NEBNext FFPE DNA Repair Mix, following the manufacturer’s protocol. To see if the repair had much effect, we left aliquots of the 2009 silica and herbarium DNA extractions unrepaired for separate library preparation.
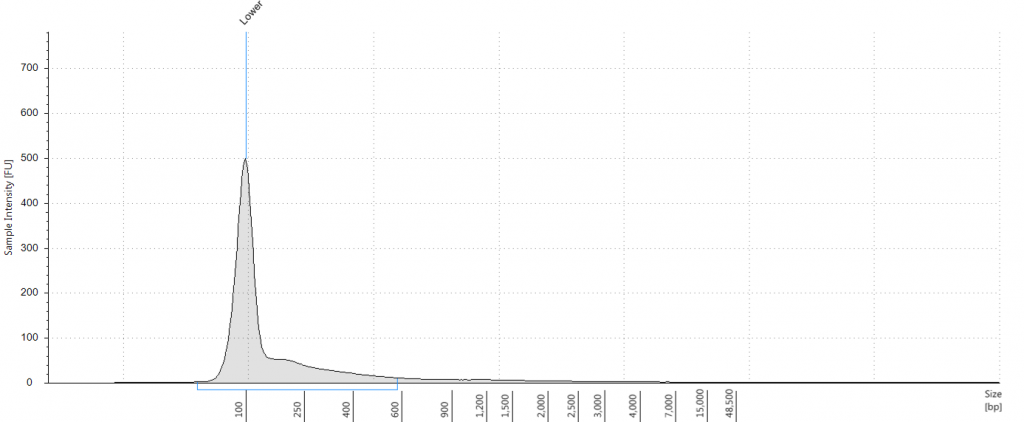
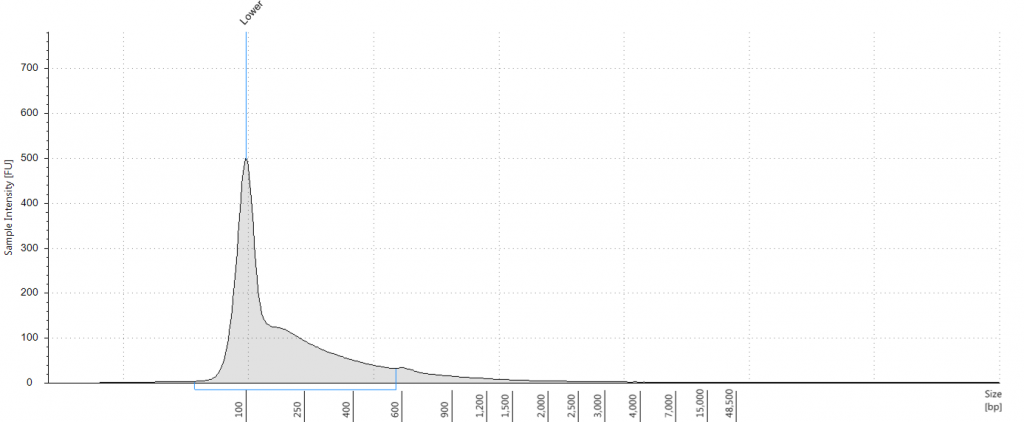
Exemplars of repaired DNA samples were run on a BioAnalyser in order to get a more accurate estimate of the degraded DNA fragment size curves. One sample (Lawrance 1932 flower) was divided into separate aliquots (a,b,c) that could be used to test different library preparation methods downstream. Unrepaired samples, and three of the repaired samples, were also run on a Tapestation, as we hoped to obtain DNA Integrity Numbers (DINs) for our samples.
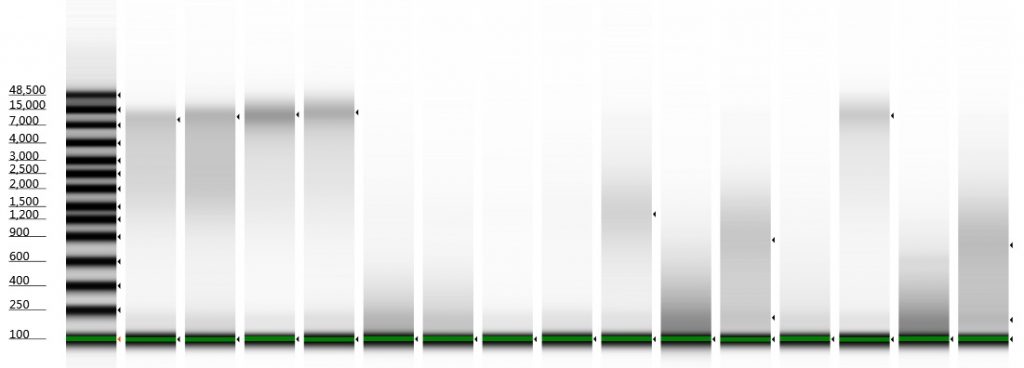
Tapestation gel of Inga umbellifera DNA; First 12 lanes – unrepaired DNA: 2009 herbarium; 2009 herbarium; 2009 silica; 2009 silica; 1948; 1840; 1840; 2004; 1932 flower; 1932 leaf; 1835. Last 3 lanes – repaired DNA: 2009 silica; 1932 flower; 1932 leaf. Green bar = 100 bp size standard present in each lane.
1. 2009 herbarium DNeasy
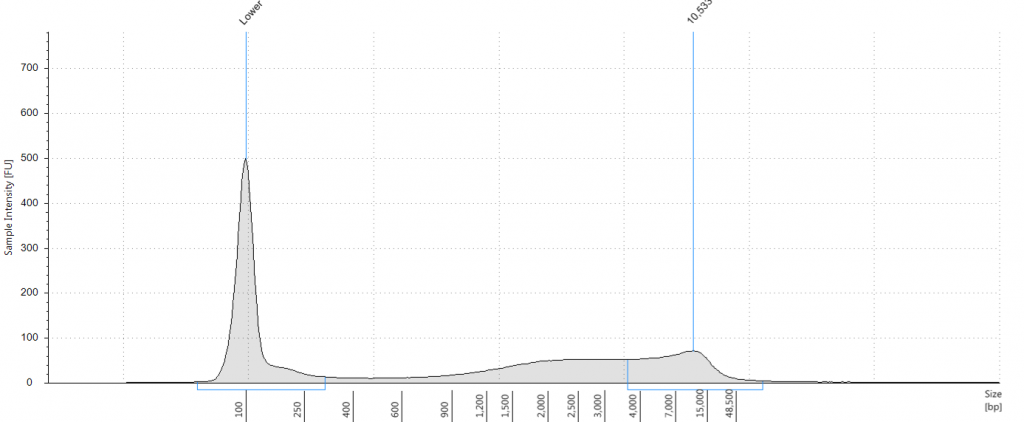
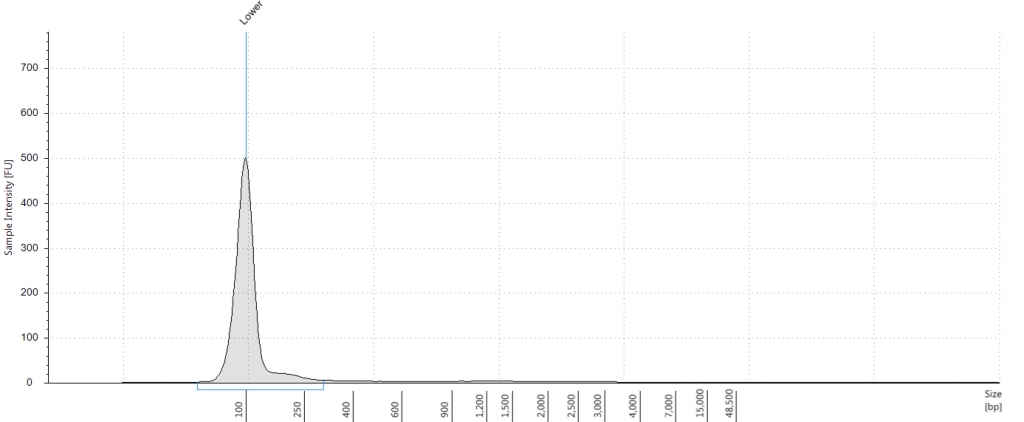
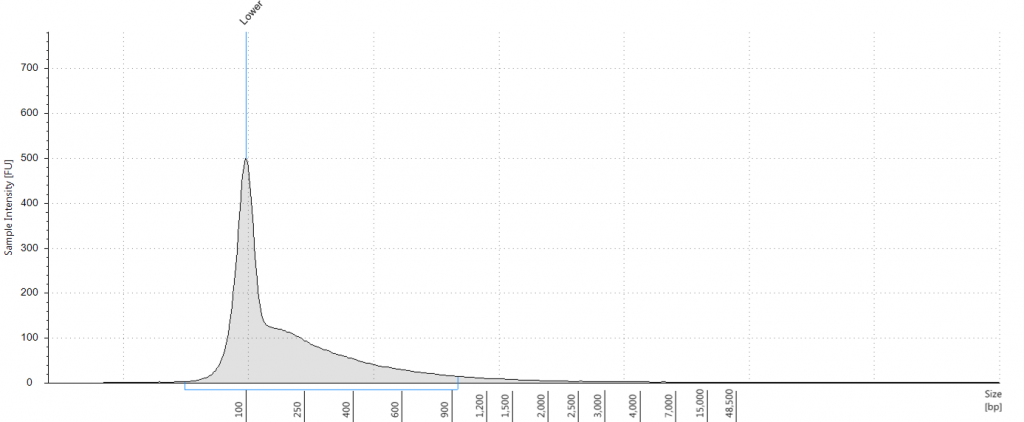
DIN 5.3; fragment size distribution 3,171 to >60,000 bp. Qubit concentration 7.88 ng/μl
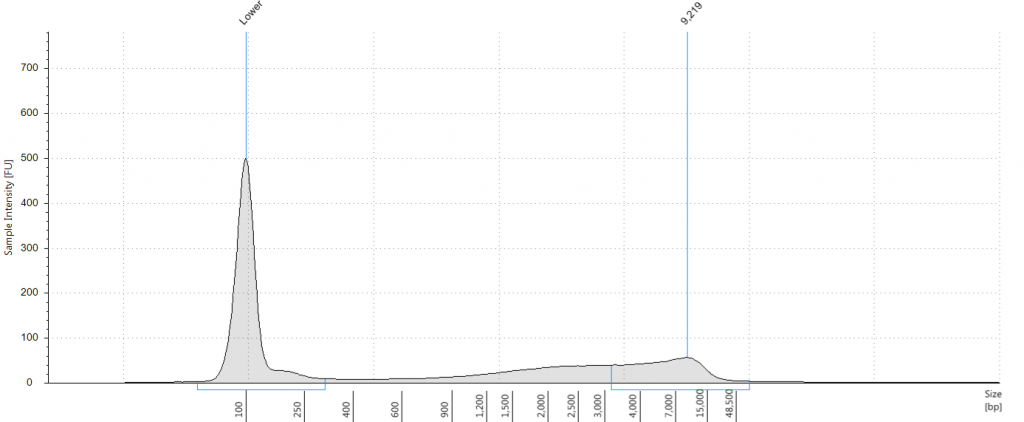
Tapestation: Inga umbellifera DNA from 2009 herbarium sheet – standard extraction (peak at 100 bp is the lane standard)
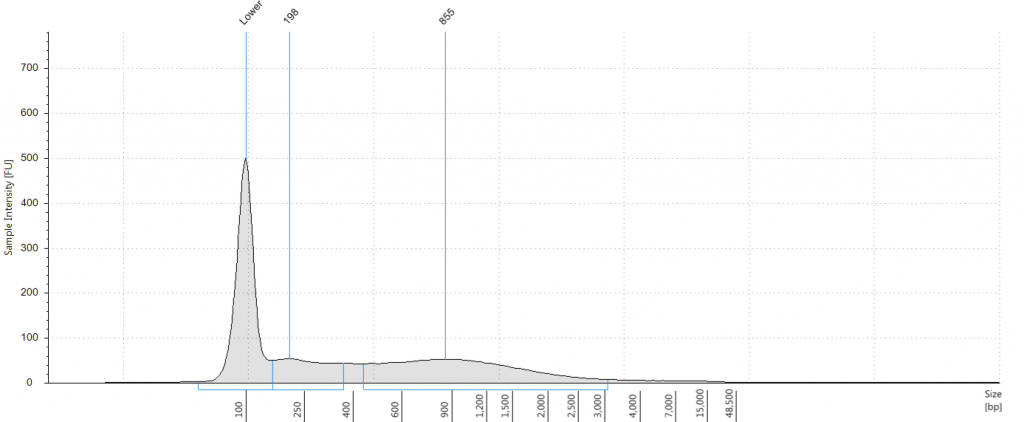
2. 2009 herbarium QiaQuick
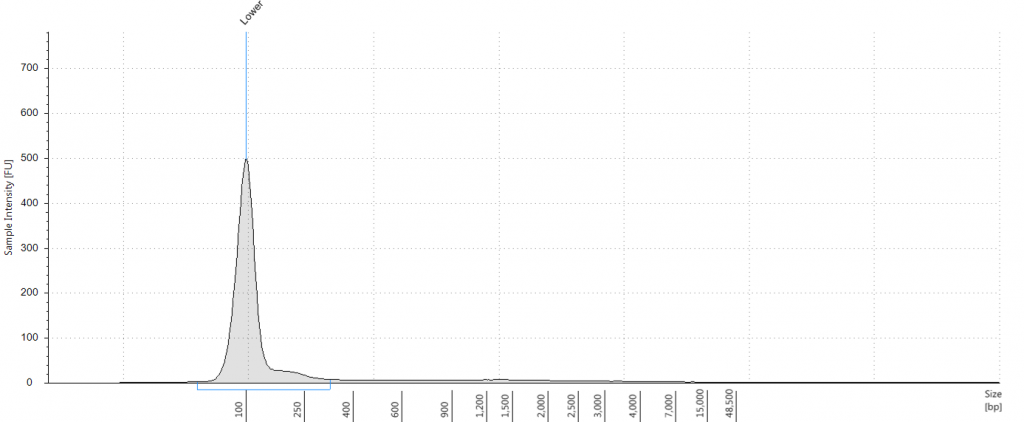
DIN 5.3; fragment size distribution 3,626 to >60,000 bp. Qubit concentration 11.5 ng/μl
3. 2009 silica DNeasy
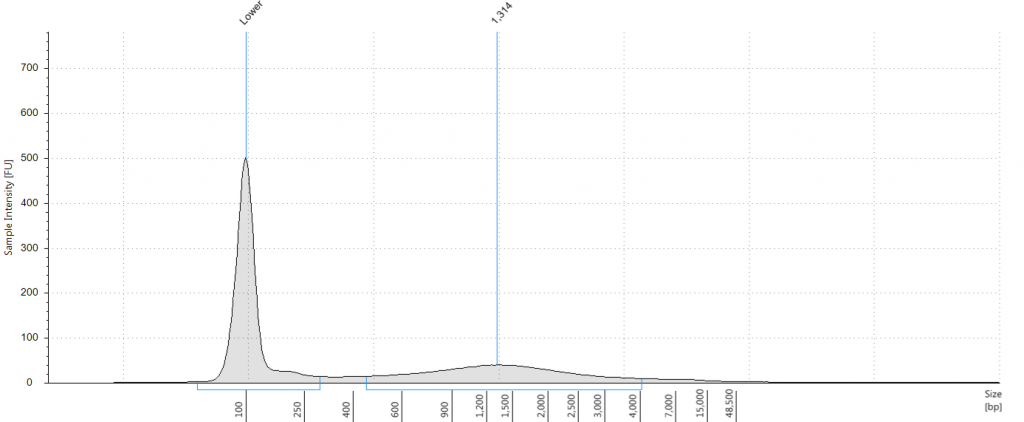
DIN 6.3; fragment size distribution 2,720 to >60,000 bp. Qubit concentration 10.8 ng/μl
4. 2009 silica QiaQuick
DIN 6.2; fragment size distribution 2,811 to >60,000 bp. Qubit concentration 8.6 ng/μl
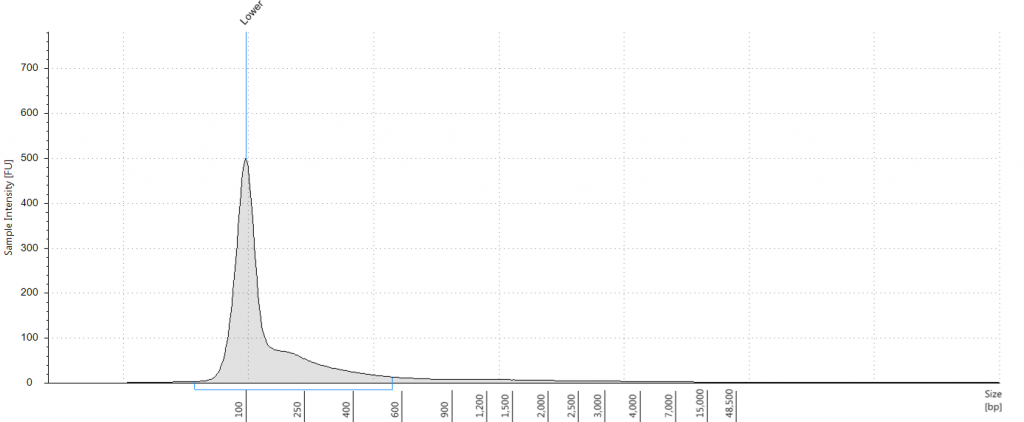
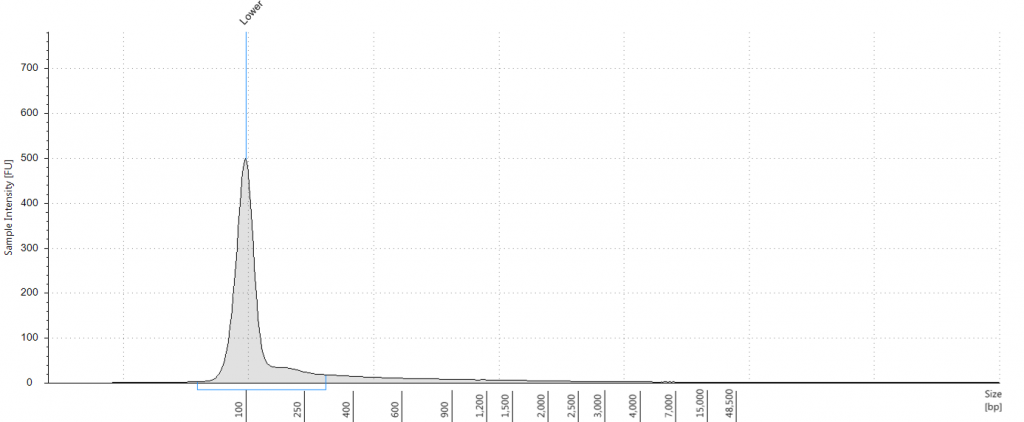
5. 1948 DNeasy
DIN not given; fragment size distribution 44 to 552 bp. Qubit concentration 2.8 ng/μl
6. 1948 DNeasy
DIN not given; fragment size distribution 47 to 576 bp. Qubit concentration 1.92 ng/μl
7. 1840 QiaQuick
DIN not given; fragment size distribution 46 to 302 bp. Qubit concentration 0.116 ng/μl
8. 1840 DNeasy
DIN not given; fragment size distribution 46 to 317 bp. Qubit concentration 0.098 ng/μl
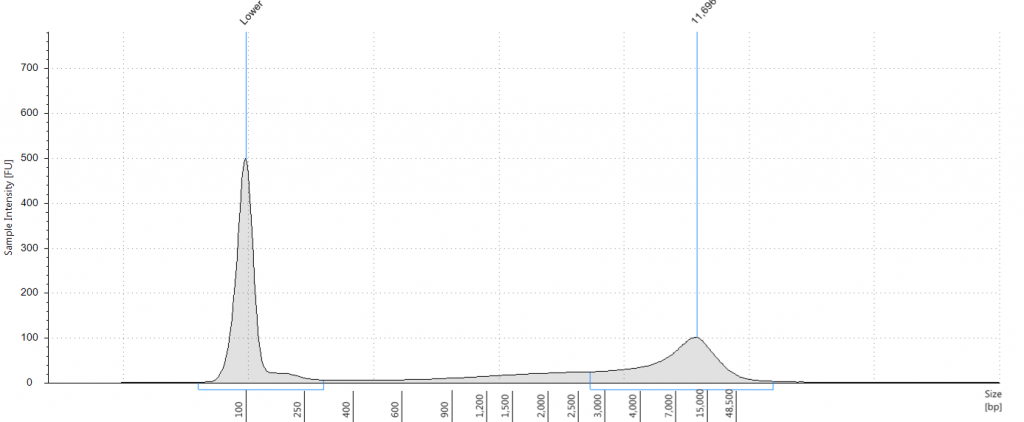
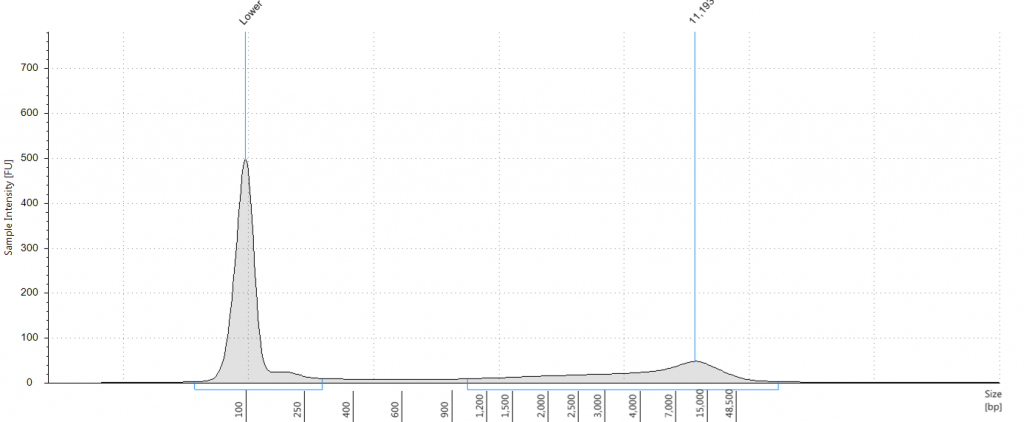
9. 2004 herbarium DNeasy
DIN 2.7; fragment size distribution 443 to 4,097 bp. Qubit concentration 4.88 ng/μl
10. 1932 flower DNeasy
DIN not given; fragment size distribution 38 to 937 bp. Qubit concentration 41.9 ng/μl
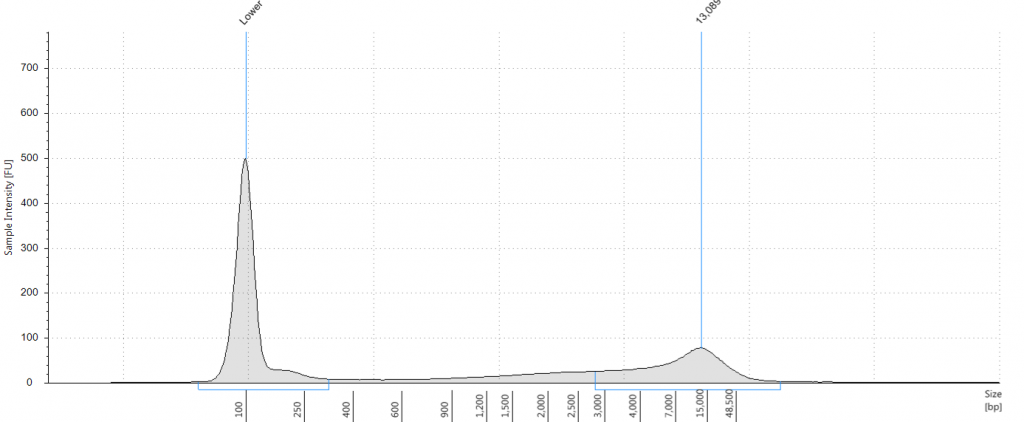
11. 1932 leaf DNeasy
DIN 1.7; fragment size distribution 48 to 3,067 bp. Qubit concentration 14.7 ng/μl
12. 1835 DNeasy
DIN not given; fragment size distribution 46 to 306 bp. Qubit concentration 1.23 ng/μl
13. 2009 silica QiaQuick with Repair Mix
DIN 6.0; fragment size distribution 1,029 to >60,000 bp.
14. 1932 flower DNeasy with Repair Mix
DIN not given; fragment size distribution 45 to 568 bp.
15. 1932 leaf DNeasy with Repair Mix
DIN 1.8; fragment size distribution 47 to 3,173 bp.
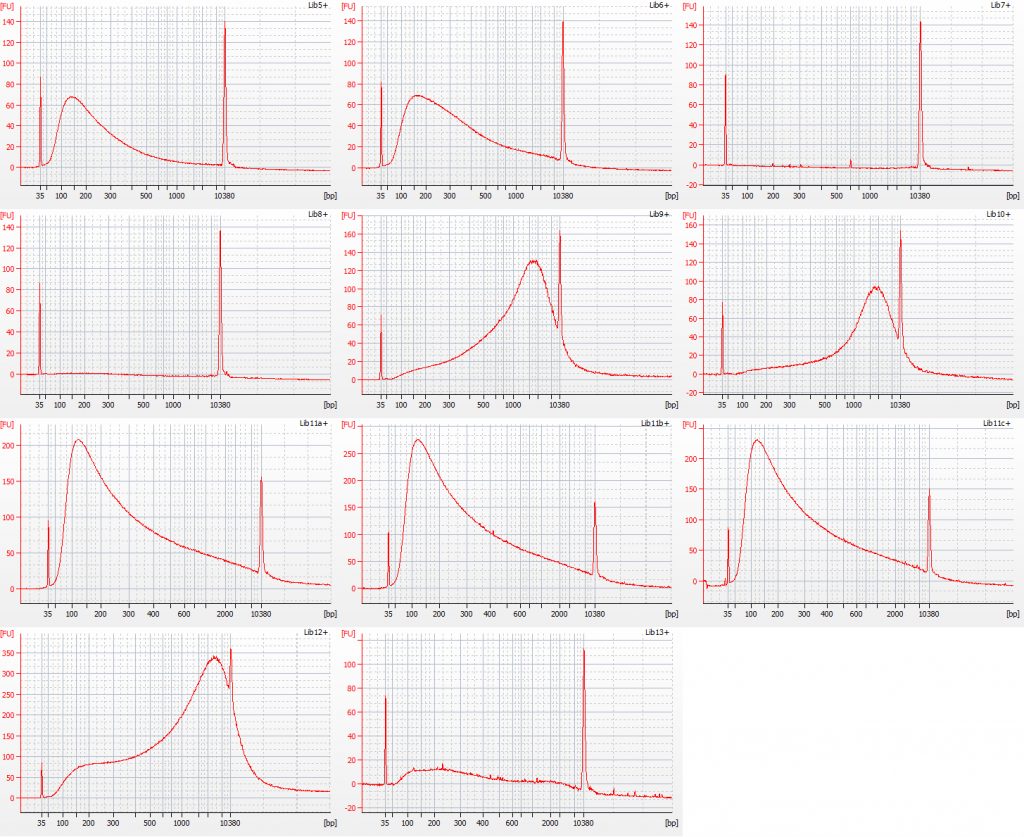
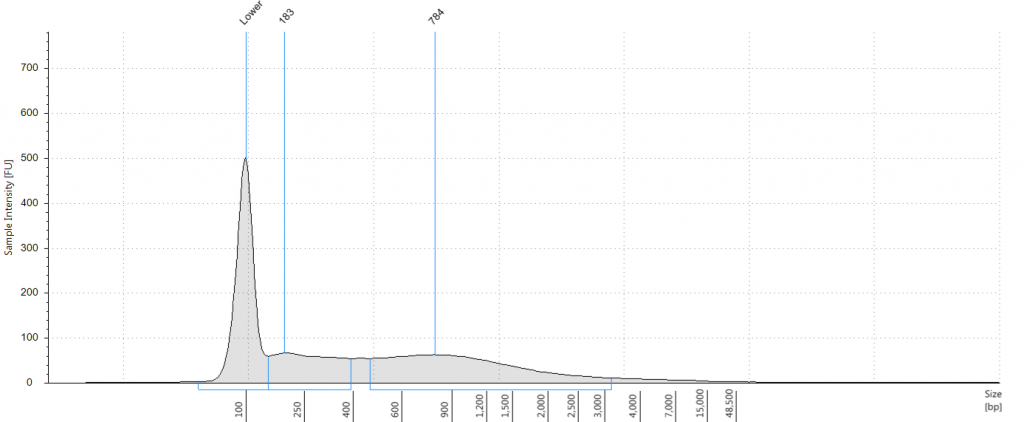
Bioanalyser traces for repaired Inga umbellifera DNA from (top left to bottom right) 1948, 1948, 1840, 1840, 2004, 2004, 1923 flower a, 1932 flower b, 1932 flower c, 1932 leaf, 1835
Unsurprisingly the highest DIN values were given to the silica-dried material from 2009 (6.2-6.3), and the second highest to the herbarium material from 2009 (5.3). The herbarium material from 2004 had the third highest DIN (2.7), but this represented a substantial drop from the 2009 collection. The only other DNA extraction that was of sufficience concentration to be assigned a DIN was the leaf extraction from 1932. These DINs were broadly similar for the unrepaired and repaired DNA, where comparisons could be made.
The amounts of DNA recovered tended to decline with the age of the sample, with one remarkable exception. All three extractions from the 1932 specimen gave very high yields, albeit of highly degraded DNA (more degraded, but higher concentration, for the flower sample; less degraded, but lower concentration, for the leaf sample). This is particularly obvious in the agarose gel image. DNA repair also caused a large drop in DNA concentration; following repair our 1840 samples had very little to no measurable DNA using the Qubit dsDNA HS kit.
Capturing Genes from Herbaria. I. What it’s all about. http://stories.rbge.org.uk/archives/16411
Capturing Genes from Herbaria. II. Inga. http://stories.rbge.org.uk/archives/16427
Capturing Genes from Herbaria. III. The samples. http://stories.rbge.org.uk/archives/16441
Capturing Genes from Herbaria. IV. DNA. http://stories.rbge.org.uk/archives/16470
Capturing Genes from Herbaria. V. Fragmenting the DNA. http://stories.rbge.org.uk/archives/16525
Capturing Genes from Herbaria. VI. Size Selection. http://stories.rbge.org.uk/archives/16645
Capturing Genes from Herbaria. VII. Comparisons. http://stories.rbge.org.uk/archives/16737
Capturing Genes from Herbaria. VIII. Amplification. http://stories.rbge.org.uk/archives/16788
Capturing Genes from Herbaria. IX. Hybrid capture. http://stories.rbge.org.uk/archives/17298
Capturing Genes from Herbaria. X. An update. http://stories.rbge.org.uk/archives/20751
Capturing Genes from Herbaria. XI. Some metagenomics of a herbarium specimen. http://stories.rbge.org.uk/archives/20817