By mid-May 2015, we had 32 separate Inga umbellifera libraries, 15 generated using the Illumina Tru-Seq Nano library preparation kits, and 17 with the NEBNext Ultra library preparation kit for Illumina. Each of these libraries had DNA index sequence incorporated that would allow the final reads to be bioinformatically separated. A single-index protocol was used; for the TruSeq libraries, we used the index set from Nicholls et al. (2015), while for the NEB libraries we had a set of 24 indices (NEBNext Multiplex Oligos for Illumina – index primers set 1; 24 reactions (E7335S)). We knew that these were going to be run on three lanes of Illumina MiSeq, for 250 base pairs of paired-end reads, so we needed to separate our libraries into three pools.
We sorted all 32 Tru-Seq and NEB post-amplification libraries into three pools of libraries with compatible index sequences, taking into account average library fragment size, for three separate hybrid capture reactions. One pool comprised the “higher quality” libraries with larger average fragment lengths, one comprised the “lower quality” libraries with shorter average fragment lengths, and the third was made up of samples that fell in between the two. The DNA sequences of the 6-base pair indices for each pool were compared to check that there was sufficient variability at all base positions for the MiSeq run to work properly.
The libraries were diluted to 10nM, and equimolar amounts were added into the three pools (15 μl per library for pool 1, and 18 μl per library for pools 2 and 3).
Pool 1 (yellow): 10 libraries: 473-649 bp: no DNA repair: Tru-Seq and NEB, 2009 herbarium and 2009 silica; DNA repair: Tru-Seq and NEB, 2009 herbarium and 2009 silica.
Pool 2 (pink): 9 libraries: 347-619 bp: no DNA repair: Tru-Seq and NEB, 2009 herbarium; DNA repair: Tru-Seq and NEB, 2004 herbarium, 1932 leaf; Tru-Seq, 1932 flower (size select 2).
Pool 3 (green): 13 libraries: 279-419 bp: DNA repair: TruSeq and NEB, 1948, 1932 flower, 1835; NEB, 1840, 1932 flower (no size select).
The total volumes were then concentrated down to 6.5 μl for each pool, in a SpeedVac vacuum concentrator.
| Year | Sample | DNA repair | Sonic. cycles | Post-sonic cleanup | Size Select | ng | Library no. | ampl. cycles | Pool | MiSeq lane |
| 2009 | H-L | – | 8 | yes | normal | 100 | NEB1- | 7 | pink | 1/9 |
| 2009 | H-L | – | 8 | yes | normal | 101 | TS1- | 8 | pink | 1/9 |
| 2009 | H-L | + | 8 | yes | normal | 101.5 | NEB1+ | 7 | yellow | 1/10 |
| 2009 | H-L | + | 8 | yes | normal | 101 | TS1+ | 8 | yellow | 1/10 |
| 2009 | H-L | – | 8 | yes | normal | 100 | NEB2- | 7 | yellow | 1/10 |
| 2009 | H-L | – | 8 | yes | normal | 101 | TS2- | 8 | yellow | 1/10 |
| 2009 | H-L | + | 8 | yes | normal | 28.2 | NEB2+ | 10 | yellow | 1/10 |
| 2009 | H-L | + | 8 | yes | normal | 70.6 | TS2+ | 9 | yellow | 1/10 |
| 2009 | S-L | – | 8 | yes | normal | 100 | NEB3- | 7 | yellow | 1/10 |
| 2009 | S-L | – | 8 | yes | normal | 101 | TS3- | 8 | yellow | 1/10 |
| 2009 | S-L | + | 8 | yes | normal | 94 | NEB3+ | 7 | yellow | 1/10 |
| 2009 | S-L | + | 8 | yes | normal | 101 | TS3+ | 8 | yellow | 1/10 |
| 1948 | H-L | + | — | — | modified | 22.9 | NEB5+ | 10 | green | 1/13 |
| 1948 | H-L | + | — | — | modified | 70 | TS5+ | 8 | green | 1/13 |
| 1948 | H-L | + | — | — | modified | 58.8 | NEB6+ | 7 | green | 1/13 |
| 1948 | H-L | + | — | — | modified | 80 | TS6+ | 8 | green | 1/13 |
| 1840 | H-L | + | — | — | — | very low | NEB7+ | 12 | green | 1/13 |
| 1840 | H-L | + | — | — | — | 5 | NEB8+ | 12 | green | 1/13 |
| 2004 | H-L | + | 3 | yes | normal | 100 | NEB9+ | 7 | pink | 1/9 |
| 2004 | H-L | + | 3 | yes | normal | 101 | TS9+ | 8 | pink | 1/9 |
| 2004 | H-L | + | 3 | yes | normal | 39.1 | NEB10+ | 10 | pink | 1/9 |
| 2004 | H-L | + | 3 | yes | normal | 70.6 | TS10+ | 9 | pink | 1/9 |
| 1932 | H-F | + | — | — | modified | 80 | NEB11+a | 8 | green | 1/13 |
| 1932 | H-F | + | — | — | modified | 80 | TS11+a | 8 | green | 1/13 |
| 1932 | H-F | + | — | — | modified | 80 | NEB11+b | 8 | green | 1/13 |
| 1932 | H-F | + | — | — | modified | 80 | TS11+b | 8 | green | 1/13 |
| 1932 | H-F | + | — | — | modified | 40 | TS11+b2 | 8 | pink | 1/9 |
| 1932 | H-F | + | — | — | — | 40 | NEB11+bv2 | 10 | green | 1/13 |
| 1932 | H-L | + | 3 | yes | normal | 100 | NEB12+ | 7 | pink | 1/9 |
| 1932 | H-L | + | 3 | yes | normal | 101 | TS12+ | 8 | pink | 1/9 |
| 1835 | H-L | + | 1 | no | modified | 16 | NEB13+ | 10 | green | 1/13 |
| 1835 | H-L | + | 1 | no | modified | 40.4 | TS13+ | 10 | green | 1/13 |
The hybrid baits that were used had been generated for an earlier project (Nicholls et al. 2015); they covered 276 target loci comprising 907 putative exons, with 3x tiling of 120 base pair RNA baits for each target exon.
Hybrid enrichment was performed on the three pools, using a single capture reaction per pool, following the MYbaits v2.3.1 protocol, with 18.5 hours of hybridisation, a high stringency post-hybridisation wash, and a final amplification using Herculase II Fusion polymerase. Instead of blocking with salmon sperm or a human block, we used adaptor block 3 for a single index kit. During the final clean up, using Qiagen PCR purification columns, we eluted in 30 μl EB, let sit for 3 minutes, eluted, then reapplied the elute and let sit for a second 3 minutes before re-eluting. We tested both 13 cycles and 14 cycles of post-hybridisation PCR, to see which gave the better post-capture libraries. All six post-capture libraries were quantified using Qubit and their size distributions assessed on a Bioanalyser.
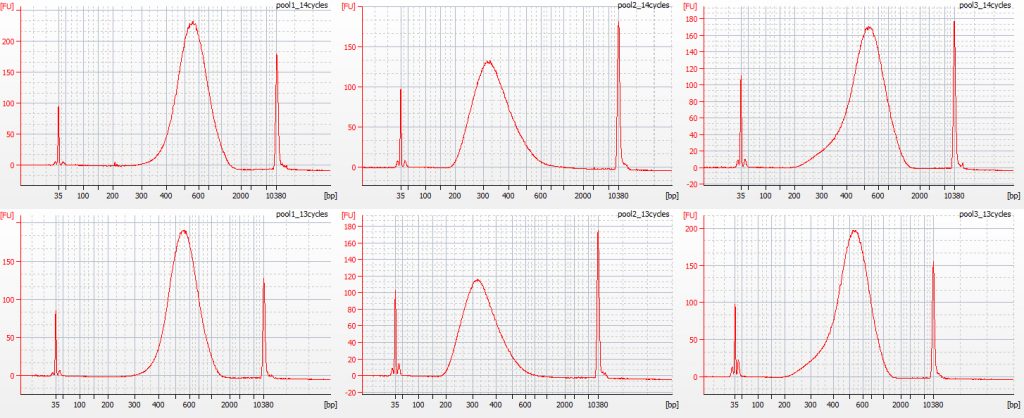
Bioanalyser traces for the six post-capture libraries – Pool 1, 2 and 3; with 14 PCR cycles (above) and 13 PCR cycles (below)
The Bioanalyser traces for the two different amplifications for each pool are reassuringly similar. On the basis of the Qubit results, the libraries that had undergone 13 cycles of PCR contained from 2.44-3.78 ng/μl DNA, while the libraries that had undergone 14 cycles of PCR contained from 6.28-12.3 ng/μl DNA.
The libraries generated with 14 cycles of PCR were thus the ones that were diluted to 10 nM, and 35 μl of each was submitted to Edinburgh Genomics for sequencing on the 15th of May 2015.
James A. Nicholls, R. Toby Pennington, Erik J.M. Koenen, Colin E. Hughes, Jack Hearn, Lynsey Bunnefeld, Kyle G. Dexter, Graham N. Stone & Catherine A. Kidner. 2015. Using targeted enrichment of nuclear genes to increase phylogenetic resolution in the neotropical rain forest genus Inga (Leguminosae: Mimosoideae). Frontiers in Plant Science 6: 710. doi: 10.3389/fpls.2015.00710
Capturing Genes from Herbaria. I. What it’s all about. http://stories.rbge.org.uk/archives/16411
Capturing Genes from Herbaria. II. Inga. http://stories.rbge.org.uk/archives/16427
Capturing Genes from Herbaria. III. The samples. http://stories.rbge.org.uk/archives/16441
Capturing Genes from Herbaria. IV. DNA. http://stories.rbge.org.uk/archives/16470
Capturing Genes from Herbaria. V. Fragmenting the DNA. http://stories.rbge.org.uk/archives/16525
Capturing Genes from Herbaria. VI. Size Selection. http://stories.rbge.org.uk/archives/16645
Capturing Genes from Herbaria. VII. Comparisons. http://stories.rbge.org.uk/archives/16737
Capturing Genes from Herbaria. VIII. Amplification. http://stories.rbge.org.uk/archives/16788
Capturing Genes from Herbaria. IX. Hybrid capture. http://stories.rbge.org.uk/archives/17298
Capturing Genes from Herbaria. X. An update. http://stories.rbge.org.uk/archives/20751
Capturing Genes from Herbaria. XI. Some metagenomics of a herbarium specimen. http://stories.rbge.org.uk/archives/20817


