There’s an exciting project, The 10KP (10,000 Plants) Genome Sequencing Project, that aims to sequence and characterize representative genomes from every major clade of embryophytes, green algae, and protists (except fungi) within the next 5 years.
Assisted by North Berwick High School pupil Heather Fortune, who has been getting work experience in the molecular labs over the summer, we took a look at how much bryophyte tissue is needed to get the sort of DNA yields needed for genome sequencing. The 10KP Genome Sequencing Project requires either > 50 μg of > 20 kb DNA fragments, or > 5 ng of > 50 kb DNA fragments. That’s high quality DNA, extracted from a single individual plant. For an oak tree or an African violet, you can pick a leaf, do an extraction, and go back for more from the same plant if you need to. With tiny plants like bryophytes it’s a lot more complicated!
We picked Conocephalum conicum as our test subject. It forms big mats all over the RBGE Fern house, and so is easy to get hold of.
(Ideally we would have worked from axenic cultures of bryophytes that had been started off as single spore isolates. This would have guaranteed that the DNA was extracted from a single individual, and should also have meant that we were just extracting plant DNA, instead of DNA from all the organisms that occur in and around the plants. Unfortunately we don’t currently have a bryophyte culture collection at RBGE.)
We took a mat of Conocephalum, and pulled it apart into smaller clumps of thallus that were obviously joined together, so that we could be certain that the DNA in our extraction came from an individual liverwort.
We then experimented a bit with ways of removing the rhizoids from the underside of the thallus – slicing with a razor, or plucking with forceps. We hoped that this would remove a lot of fungal material, as well as the organic debris that gets stuck between the rhizoids. The clump we chose to work with weighed about 0.34 g after we’d removed most of its rhizoids and blotted it dry.
We added about 15 mls of CTAB buffer to the liverwort powder, and split the extraction more or less equally between 16 eppendorf tubes.
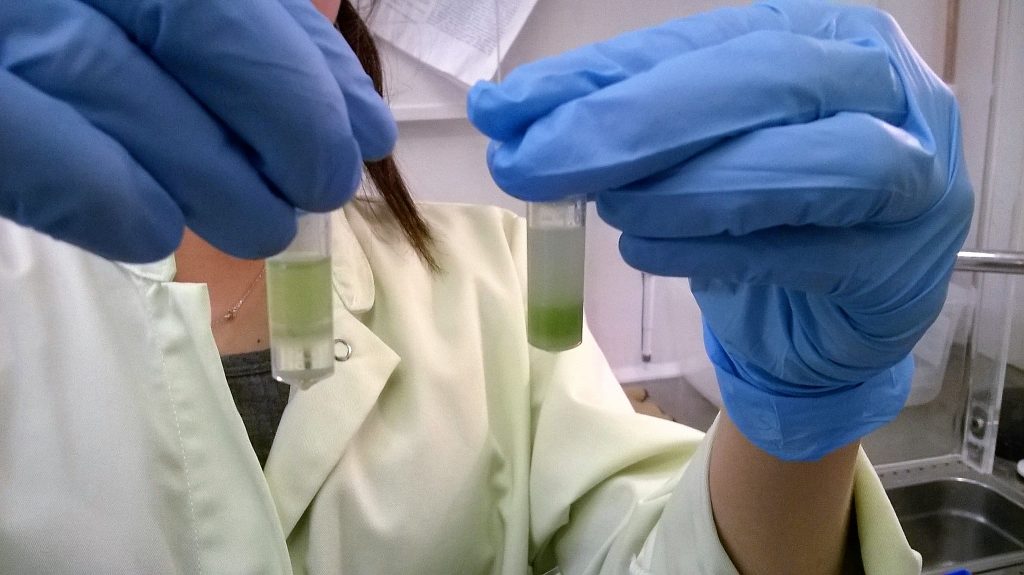 Washing the DNA extractions with chloroform:IAA (before shaking – left hand tube; after shaking – right hand tube)
Washing the DNA extractions with chloroform:IAA (before shaking – left hand tube; after shaking – right hand tube)
We precipitated the DNA overnight at -20°c using isopropanol, and resuspended each pellet in 50 μl of a standard Tris-EDTA buffer, giving us a total of 800 μl.
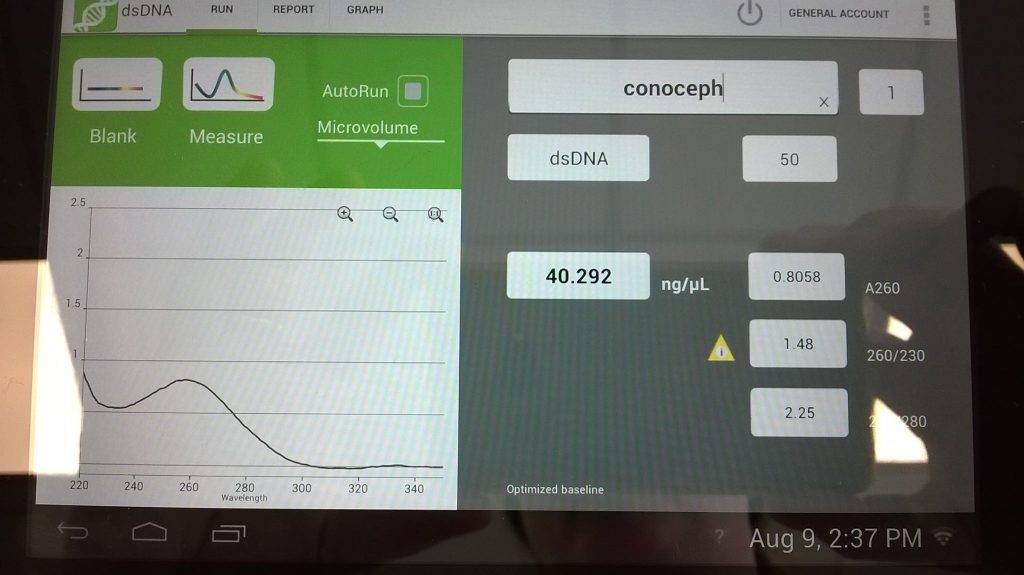
We looked at spectrophotometry and fluorometry-based quantifications using our deNovix, to assess DNA quality and quantity.
We also ran 5 μl of the DNA out on an agarose gel, to check if we had high molecular weight DNA.

Conocephalum genomic DNA on a 2% agarose TBE gel, stained with SYBRsafe, with 3.5 ul of invitrogen 1kb ladder as a size standard
In total, we have extracted about 1.86 μg of DNA from about 0.34 g of Conocephalum thallus tissue. The required > 50 μg of > 20 kb DNA fragments is about 30 times this amount! Without increasing the efficiency of the DNA extraction method, we would need nearly 10 g of the liverwort thallus. For uncultured Conocephalum, that’s a problem because we may not be able to find a patch of thallus of that size that’s obviously a single interconnected individual.