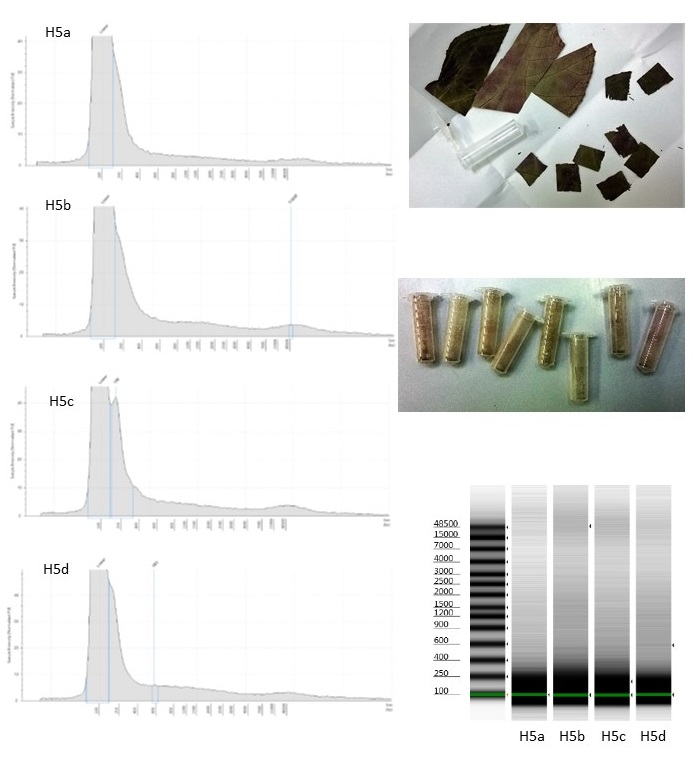
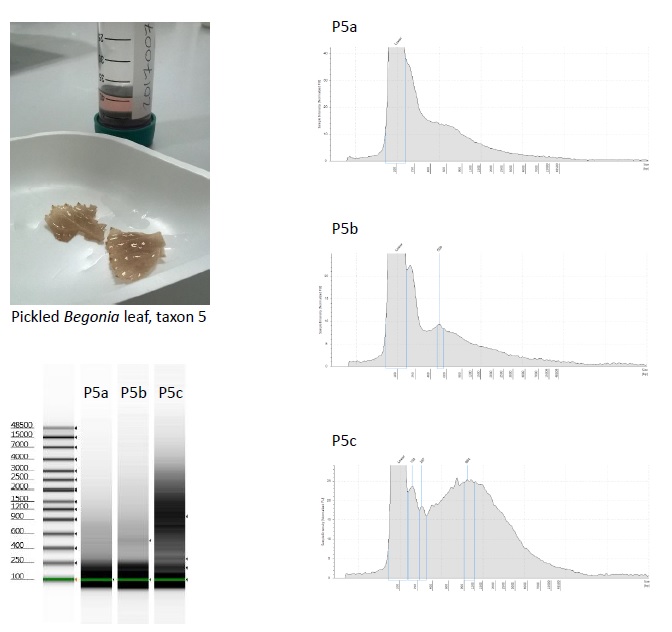
For one of the taxa in our study set, Begonia scottii (living collection no. 20170076), we made a few replicate DNA extractions using Qiagen DNeasy plant mini-kits, and assessed the DNA from the replicates.
We used plant tissue from two of the types of preservation we’d been testing – hairdryer-dried leaves, and leaves that were pickled in 70% ethanol. For the hairdryer-dried leaves, we ground the tissue in four separate eppendorf tubes, using flattened tungsten beads in a TissueLyer. We then added Qiagen’s lysis buffer to each of the tubes separately before incubating for an hour at 65°C. For the pickled leaves, tissue was first blotted dry and then washed twice in Qiagen’s lysis buffer, before being ground in the buffer using a pestle and mortar, with a little acid-washed sand. The lysis buffer was then pipetted into three separate eppendorf tubes, and incubated for an hour at 65°C.
Samples from each of the individual DNA extractions were run on an Agilent Genomic DNA ScreenTape. None of the samples were assigned a DNA Integrity Number (DIN) by the software.
For the hairdryer-dried tissue, the four extractions had comparable Tapestation profiles, with a little high molecular weight DNA apparent, but also similar quantities of degraded DNA.
The Tapestation images of the DNA extracted from the pickled material are more surprising, given that the plant tissue was all ground in a single mortar, from which the lysis buffer was aliquotted out: The fragment size distribution is different for each extraction, with a larger quantity of higher molecular weight DNA in the third extraction.
The main difference between the three DNA extractions is that the third was the last to be aliquotted, and so contained more of the “dregs” of the lysis buffer/plant tissue mixture.
The moral may be that the pickled plant tissue should have been homogenized rather better, and that higher molecular weight DNA was present in, and extracted from, small fragments of the tougher vascular tissues that ended up disproportionately in the third tube.
However, it does also mean that comparisons of metrics (e.g. average DNA fragment size, or DNA concentration) from small numbers of DNA extractions between different types of samples, as estimators of DNA preservation quality, need to be treated rather cautiously.