Following on from the rather unpredictable results we obtained from fragmenting duplicate aliquots of CTAB-extracted Polytrichum DNA in the Bioruptor, Isuru cleaned aliquots of IK31 and IK53 using Qiagen DNeasy plant mini-kits, and qubited the cleaned DNA to see if we still had enough to experiment with. Just running the two extractions through the DNeasy kit resulted in the loss of around 90% of the CTAB-extracted DNA, suggesting that some optimisation of the protocol would be wise if this step is going to be required for many of the samples needed for Isuru’s PhD work.
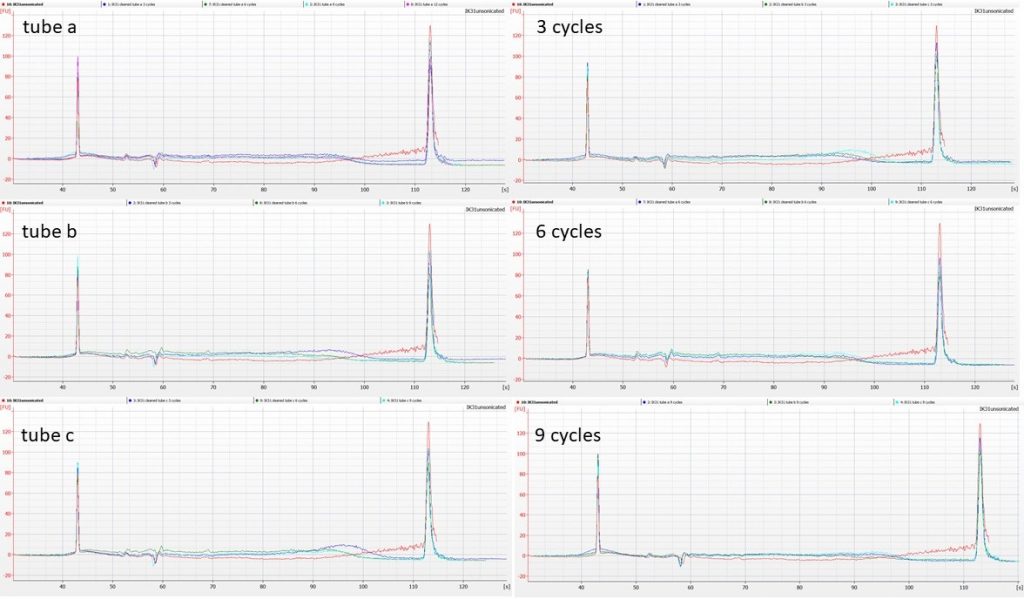
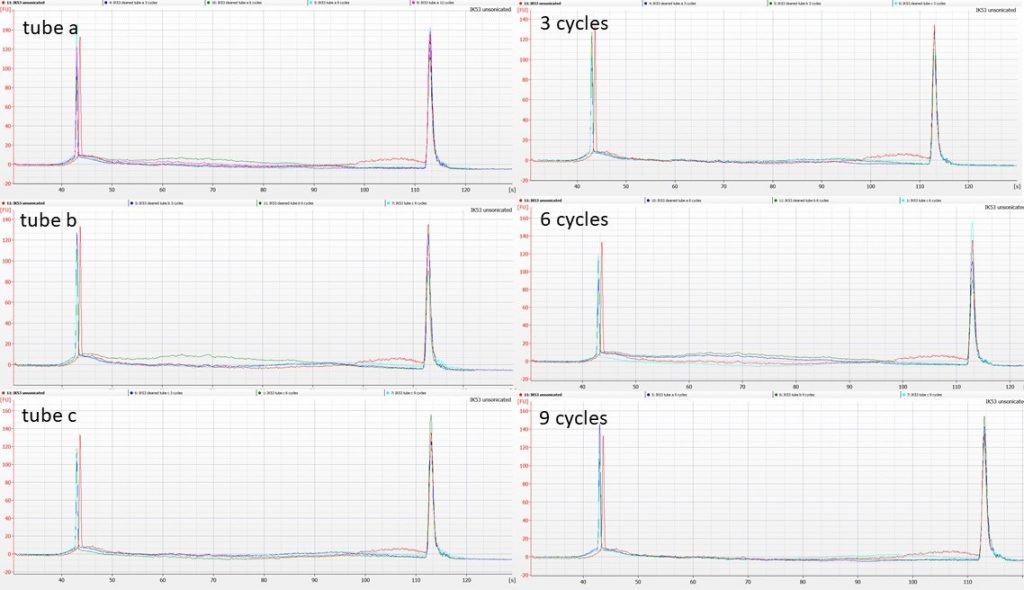
We split the cleaned Polytrichum DNA into three 53 µl aliquots per accession, and put each of them through 12 cycles of sonication (30 sec on, 90 sec off). We removed 1.25 µl from each tube after 3, 6 and 9 cycles, and ran the sonicated DNA on Agilent High Sensitivity Bioanalyser chips.
Unfortunately, the quantities of DNA are so low that patterns are a little hard to distinguish. The following illustrations show the sonication patterns per tube on the left hand side (with unsonicated DNA in red), and the patterns per number of sonication cycles on the right (again, unsonicated DNA in red), for both accessions.
In both cases, the pattern appears to be that the higher-molecular-weight DNA in the original extraction, shown as the red line, is no longer present after 3 cycles of sonication; between 3, 6, 9 and 12 cycles there’s little additional change apparent.
As far as our protocol development goes, it seems that we can stick all our samples through 2-3 cycles of sonication without worrying much about over-fragmentation. We do have a decision to make about whether or not to clean the DNA, and if so, how. That will have to wait until we’ve had a chance to compare how the DNA performs during library prep, and consider whether having a far larger quantity of input DNA (and so, for example, being able to size select) outweighs any issues of lower DNA quality.