
Fig. 1. Inga umbellifera, Mathews 1593, collected in Peru, Departamento San Martin: Provincia San Martin, Tarapoto, in 1835.
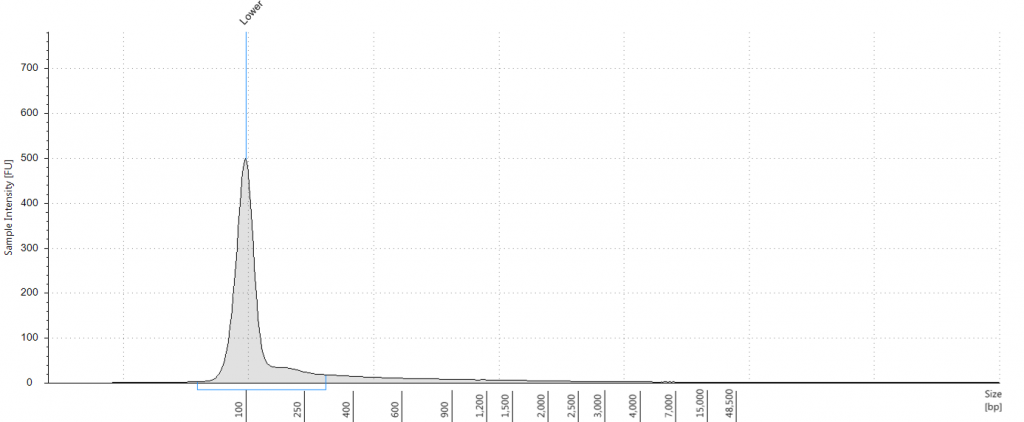
As part of our hybrid capture project, we sampled from an Inga umbellifera specimen that was collected about 180 years ago, by Andrew Mathews, in Peru in 1835. We made our Qiagen Plant DNeasy DNA extractions from the piece of plant tissue shown in Fig. 1. The DNA was very degraded, and present in quite low quantities (Tapestation fragment size distribution 46 to 306 bp; Qubit concentration 1.23 ng/μl; Fig. 2); from this we were still able to generate both Illumina Tru-Seq and NEB-Next DNA libraries (#13) that we sent to Edinburgh Genomics sequencing facility at the University of Edinburgh for Illumina Mi-Seq sequencing.
We analysed the sequence data from the Tru-Seq and NEB-Next libraries separately, as our study aimed to compare different DNA extraction and library preparation techniques; from each library, we obtained over 300 thousand bases of Inga DNA sequence data that matched to our hybrid bait set. For the two libraries combined, we obtained over 1.6 million reads that passed quality controls, over 85% of which matched the hybrid baits that we’d used, and another c. 5% of which matched the Inga plastid genome. However, that still leaves around 10% of the reads that were not from the legume hybrid baits that we’d used, and we were interested to see what else might be present in that data set.
| Library | No. of trimmed reads | % reads aligned to baits | % reads aligned to Inga plastid | Average quality score of variant positions (AQV) | Number of variant bases | Loci recovered (max 276) | Conservatively called sequence (CCS), bp |
| H1835_NEB13+ | 1,013,414 | 87.40% | 4.30% | 139.18 | 7,186 | 249 | 317,244 |
| H1835_Tru13+ | 659,161 | 84.20% | 5.20% | 132.97 | 7,045 | 247 | 310,949 |
We compared the DNA sequences of the reads from one of these libraries, H1835_NEB13+, to a publically available database (using a blast analysis run using Galaxy). This gave us a metagenomic analysis of the H1835_NEB13+ library reads, so that we could tell if anything other than Inga DNA had been in the extractions. Of 12,754,803 untrimmed reads, a huge number, over 8.3 million, were matches to legume sequences, exactly as we would expect from an Inga specimen (65%, with 1,211,319 Ingeae tribe reads, 2,015,461 Mimosioid legume reads, and 5,088,509 Fabaceae reads).
But not all the sequence reads were matches to plant DNA…
There were also:
3,047 reads that matched Homo sapiens (0.02%, which may or may not come from our intrepid plant collector, Mr Andrew Mathews, from his Peruvian wife, from the herbarium worker who stuck the pressed plant onto the card sheet, from taxonomists who have handled the specimen over the years, or indeed from anyone in the laboratory when the DNA extractions were being made…),
88,216 reads that matched Mediterannean mussels (0.7%),
and 346,820 reads that matched Streptococcus (2.7%; always wash your hands after touching herbarium specimens!).
Humans and bacteria are pretty easy to explain – people have been handing this plant material ever since it was collected, and there are bugs everywhere. But what about the mussels? The city of Tarapota in Peru is a long way from the coast, but after the plant was collected, it was squashed and dried in a plant press, and transported across the country, and eventually over to Europe. When we first got these results, we imagined working dinners in the Mathews household, with the botanist splitting his attention between his plant collections and a towering plateful of shellfish, dripping mollusc juices across the specimens. It does, however, seem unlikely that a professional plant collector would be quite that careless.
An alternative explanation is that we’ve also extracted DNA from glue. The leaf material we are working with has been removed from a herbarium sheet, to which it had been stuck. Animal-based glues were common in the 19th century, and although the classic glues were from mammal hides and fish, mussels certainly have a lot of sticky potential.
References:
Hart, M.L., L.L. Forrest, J.A. Nicholls & C.A. Kidner. In press. Retrival of hundreds of nuclear loci from herbarium specimens. Taxon.
James A. Nicholls, R. Toby Pennington, Erik J.M. Koenen, Colin E. Hughes, Jack Hearn, Lynsey Bunnefeld, Kyle G. Dexter, Graham N. Stone & Catherine A. Kidner. 2015. Using targeted enrichment of nuclear genes to increase phylogenetic resolution in the neotropical rain forest genus Inga (Leguminosae: Mimosoideae). Frontiers in Plant Science 6: 710. doi: 10.3389/fpls.2015.00710
Capturing Genes from Herbaria. I. What it’s all about. http://stories.rbge.org.uk/archives/16411
Capturing Genes from Herbaria. II. Inga. http://stories.rbge.org.uk/archives/16427
Capturing Genes from Herbaria. III. The samples. http://stories.rbge.org.uk/archives/16441
Capturing Genes from Herbaria. IV. DNA. http://stories.rbge.org.uk/archives/16470
Capturing Genes from Herbaria. V. Fragmenting the DNA. http://stories.rbge.org.uk/archives/16525
Capturing Genes from Herbaria. VI. Size Selection. http://stories.rbge.org.uk/archives/16645
Capturing Genes from Herbaria. VII. Comparisons. http://stories.rbge.org.uk/archives/16737
Capturing Genes from Herbaria. VIII. Amplification. http://stories.rbge.org.uk/archives/16788
Capturing Genes from Herbaria. IX. Hybrid capture. http://stories.rbge.org.uk/archives/17298
Capturing Genes from Herbaria. X. An update. http://stories.rbge.org.uk/archives/20751
Capturing Genes from Herbaria. XI. Some metagenomics of a herbarium specimen. http://stories.rbge.org.uk/archives/20817

Cymon
Ha… interesting. Someone should do a study of herbarium preparation materials over the centuries to see what potential DNA they contain. When I was at NHM (last century), I remember being forwarded a question from a member of the public about whether DNA could be extracted from paper. I’m not sure what answer I gave now, but I think it at least possible given that a few cambium cells are certain to be in the pulp mix before processing. So it’s more a matter of what the processing removes. I would guess you certainly can get DNA of the trees species from which the paper is made; I guess someone has done this now… C.