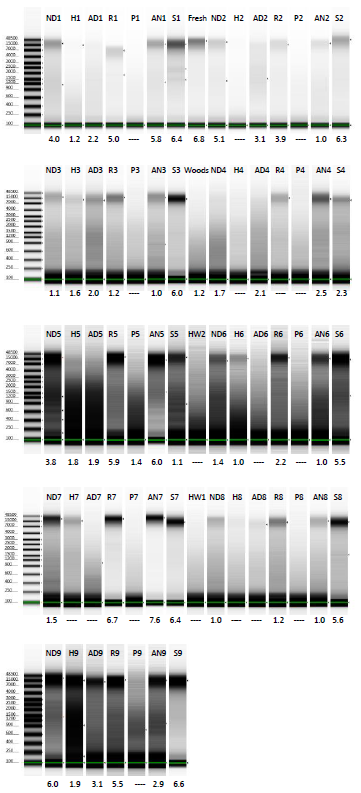
Keen to see the effects of different specimen preservation techniques on DNA quantity and quality, we have assessed extractions of DNA from nine Begonia accessions x seven preservation treatments (room temperature = AN; in the RBGE drying room = ND; in the RBGE drying room after an alcohol wash = AD; with a hairdryer = H; pickled in alcohol = P; preserved in RNAlater = R; in silica gel = S).
The DNA was extracted by Hannah Wilson, using standard Qiagen Plant DNeasy mini-kits.
The DNA extractions were quantified on our deNovix, using Qubit ds HS fluorometry kits, with 1 µl of extraction per assay, and duplicates for each DNA extraction. We also took spectrophotometry readings on the deNovix, generating 260/230 and 260/280 ratios for all the samples.
The DNA extracted from the test set of Begonia samples that we made in February has now all been run on Tapestation Genomic DNA ScreenTape, with 1 µl of DNA extraction per lane, so that we can get an idea of patterns of DNA degradation. There’s some variation sample-to-sample, although in general, the best quality DNA is from tissue stored in silca gel, and the lowest quality DNA is from tissue that’s been pickled in alcohol.