We started our lab work on the Polytrichum hybrid baits project on the 1st of October, by normalising some CTAB-extracted DNA with 0.1X TE to 55 µL of 1.905 ng/µL. The stock DNA extraction from the IK31 sample, material from New Zealand collected in 2017, had a concentration of 21.8 ng/µl with a 260/280 ratio of 2.2. The stock DNA extraction from the IK53 sample, material from Alaska collected in 2000, had a concentration of 32.6 ng/µl with a 260/280 ratio of 1.7. (Concentrations measured on a Qubit with a dsDNA HS kit, spectrophotometry readings measured on a Nanovue.)
This was followed by what was to have been a quick investigation of DNA fragmentation in the two Polytrichum accessions (one with high molecular weight DNA, IK31, and one more typical of herbarium material, IK53, with mostly degraded DNA), to devise the fragmentation protocol we would then apply across the rest of the samples in the project.
Naturally nothing is quite that simple. Firstly, possibly as a result of initially quantifying the stock DNA on a Qubit, but quantifying the normalised DNA on a DeNovix (albeit using the same Qubit dsDNA HS kit for both), the normalised DNA was far less concentrated than we expected based on the stock concentrations. And yes, we were using lo-bind tubes…
The second problem is a little more complex to describe. We were testing different numbers of sonication cycles (30 sec on, 90 sec off) in a Diagenode Bioruptor. Six 53 µl aliquots were made from the normalised DNA for both accessions, and the tubes of DNA from each accession were put in the sonicator for up to 12 cycles. After every second cycle, one tube was removed and replaced with a blank tube (containing 53 µl of 0.1 TE), i.e. for each accession, we ended up with aliquots that had been sonicated for 2, 4, 6, 8, 10 and 12 cycles.
The DNA was then run on an Agilent Genomic DNA ScreenTape…
… and on a Bioanalyser chip with a High Sensitivity DNA assay….
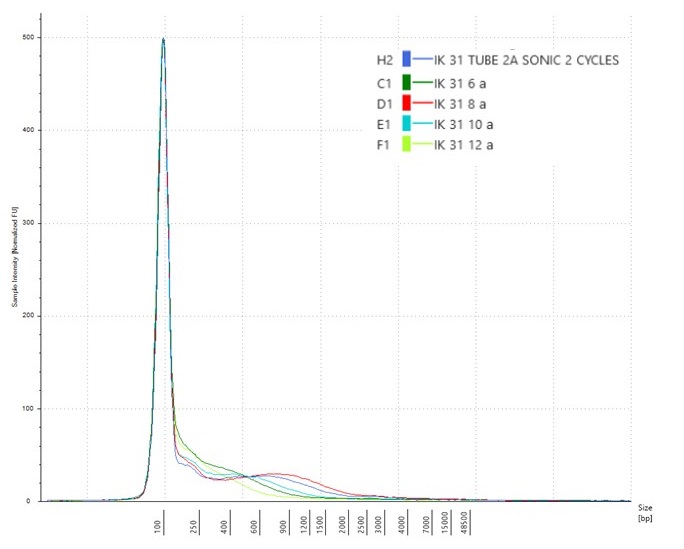
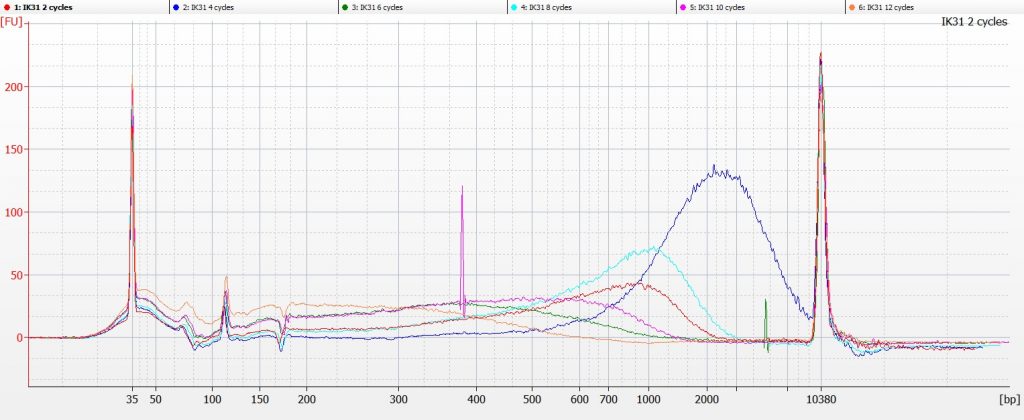
…and in both cases, the DNA fragmentation profile for IK31 suggested that there was a problem with the tube labelling, as the DNA that had been subject to 4-8 cycles of sonication contained higher molecular weight fragments than the DNA that had only had two cycles of sonication (the red line on the Bioanalyser chart above).
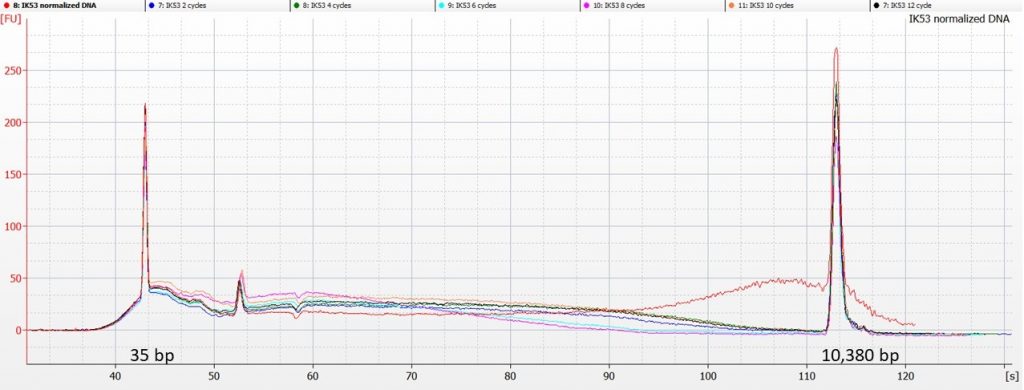
The picture for IK53 is a little more straightforward:
Here, the Bioanalyser traces show that the sonicated DNA contains less of the higher-molecular weight DNA than was present in the original aliquot (represented by the red line above). However, there’s not really a clear pattern of reducing fragment size after 2, 4, 6, 8, 10 or 12 cycles of sonication.
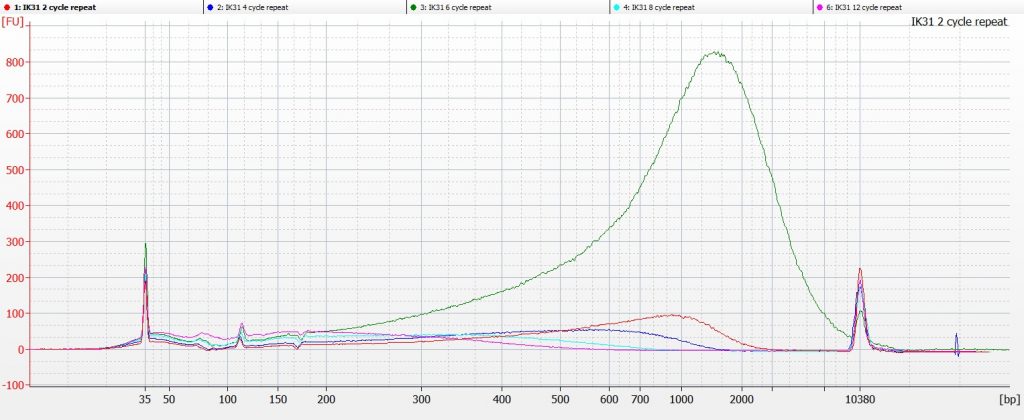
So we repeated the experiment for IK31, taking very great care to match the numbers on the tubes with the number of sonication cycles. And this time, the tube that had undergone 6 cycles of sonication (the green line on the chart below) was the one that contained the highest molecular weight DNA.
This was a bit frustrating, as this time we were even more sure that we had the right samples in the right tubes in the right wells on the Bioanalyser chip.
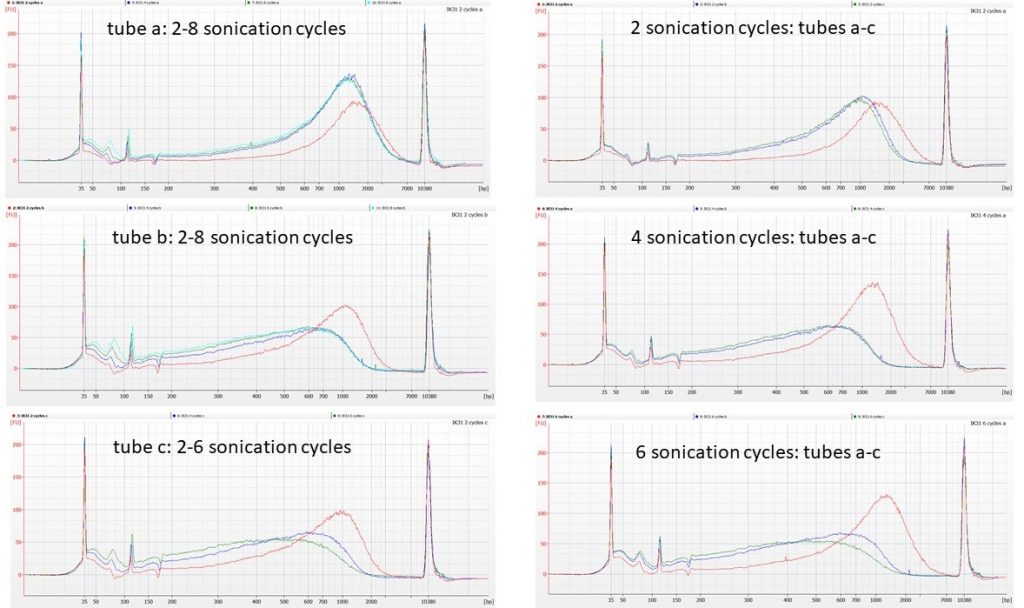
So the NEXT thing we tried was taking three (supposedly) identical aliquots of 53 µl of the normalised IK31 DNA, and putting them in the sonicator, alternating with three blank tubes that contained 53 µl of 0.1 TE. We then removed 1 µl from each of the DNA aliquots after every second sonication cycle (up to 8 sonication cycles), to run on a Bioanalyser High Sensitivity chip.
(As there are only 11 wells on these chips, the tube c 8 cycle sample was not run).
The traces seem to show that tubes b and c of the normalised DNA behave similarly to each other, in that the first four cycles of sonication remove some of the high molecular weight DNA, although between 4 cycles and 8 cycles of sonication there’s little further change in the DNA fragment size distributions. However, tube a of the normalised DNA does not appear to fragment at all, even after 8 cycles of sonication.
We’ve seen similar issues before, with DNA that is difficult to shear, particularly in plants where polysaccharides may be co-precipitating in the extractions alongside the DNA. What seems harder to explain here is that these different patterns are between aliquots from the same tube of normalised DNA.
Our next step will be to clean some of the IK31 DNA extraction and repeat the sonication experiment, to see if the results become more consistent between replicates.