After RBGE’s initial involvement in land plant DNA barcode marker selection, culminating in a couple of 2009 papers that both utilized bryophyte barcoding data sets, we started a project to DNA barcode all of the British liverworts (and hornworts).
Our much-loved staff bryologist, Dr David Long, has made a vast number of bryophyte collections over the course of his career (to date). These were to form a starting point for our study, bulked out by a mixture of targeted collecting, and an appeal for material that went out in the magazine of the British Bryological Society in 2007 (D.G. Long, P.M. Hollingsworth, L.L. Forrest. 2007. Barcoding Britain’s liverworts and hornworts: a new project and request for material. Field Bryology 93: 10-13).
It’s very hard for me to believe that 10 years have past since then, without much output from the project. This is therefore a brief update on what’s been done so far…
We now have over six thousand bryophyte DNA extractions listed in our DNA bank. However, not all of these are part of the barcoding project; some were for population biology studies (e.g., on Anastrophyllum), and others form part of phylogenetic or monographic work (e.g., Dr Sumudu Rubasinghe‘s PhD research on Cleveaceae). The British barcoding samples comprise somewhere around 2,500 accessions, representing most of our c. 300 liverwort (and hornwort) taxa. Not all of these samples are from the British Isles – where material is scarce, or taxonomic problems might be solved with other material, or we wanted to match the usage of names across a wider geographic area, we have sampled further afield.
Most of the bryophyte barcoding samples have been processed for a standard set of four plant barcode markers: three plastid loci, from rbcL, matK and psbA-trnH, and one nuclear locus, ITS2. However, sequencing success rates have varied widely, between markers and between taxonomic groups.
Laboratory work on the project has mostly been carried out by staff (past and present) from RBGE’s Scientific and Technical Services division: Dr Michelle Hart, Alexandra Clark, Ruth Hollands and myself.
We have also had a number of students and visitors working either directly on the project, or on related projects. These include:
University of Edinburgh BSc Honours, Biological Sciences:
Kenneth McKinlay (2013), DNA barcoding British liverworts: Lophocolea
RBGE / University of Edinburgh MSc, Biodiversity & Taxonomy of Plants:
David Bell (2009), DNA barcoding of the leafy liverwort genus Herbertus Gray in Europe and a review of the taxonomic status of Herbertus borealis Crundw.
Lucy Reed (2011), Barcoding British Liverworts: Plagiochila (Dumort.) Dumort.
Kimberley Fackler (2013), DNA barcoding and morphological reassessment of British species of simple thalloid liverwort genus Metzgeria
Tom Dawes (2016), Understanding the phylogenetic relationship between Anthoceros agrestis, the hornwort model organism, and the related A. punctatus using novel nuclear gene regions.
Sibbald Trust-funded visitors and projects:
David Bell (2013), DNA barcoding British mosses
Isuru Kariyawasam (2014), molecular methods and bryophyte barcoding
Dr Des Callaghan (2017), British species of the moss Weissia
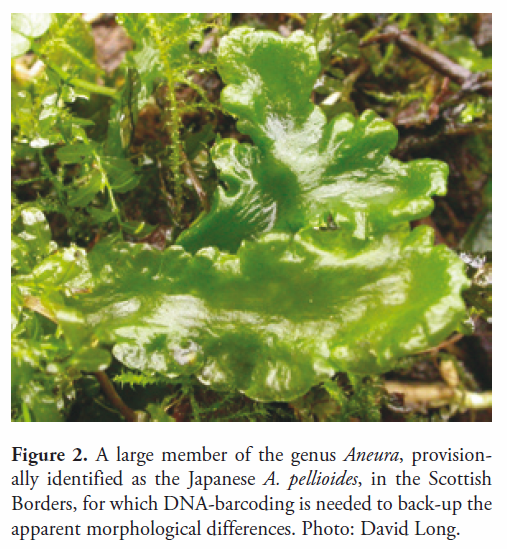
David Bell (2017), Databasing and digitizing herbarium specimens of Aneura
SYNTHESYS Access-funded visitors:
Dr Juan Carlos Villarreal Aguilar (2012), DNA barcoding European hornworts
Dr Wolfgang Hofbauer (2014, 2017), Schistidium moss in the built environment
Department for Environmental Food and Rural Affairs (DEFRA)-funded project:
David Bell (2012), The use of DNA barcoding to address major taxonomic problems for rare British bryophytes
Interns / Work experience students
Jennifer Westermann (2017), DNA barcoding liverworts
Collaborative projects: