A recent molecular project looking at species boundaries between two British Grimmia species, Grimmia donniana and Grimmia arenaria, with Dr Des Callaghan reminded me that there were still some unedited nuclear ribosomal Internal Transcribed Spacer ITS2 sequence files for the genus from Dr Wolfgang Hofbauer’s EU-SYNTHESYS-funded visit to RBGE back in 2014. At that point we had been looking into mosses that grow on building surfaces, and had ended up focusing on another genus, Schistidium, instead of Grimmia. This new project seemed like the ideal opportunity to dig out and utilize the older unused files.
However, having unearthed and edited the 2014 Grimmia sequence files, I discovered that the genetic variation between them and the species that we are currently working on is too high to make useful comparisions. The 19 sequences from 2014 were also not an exact match to any identified plants in the NCBI database, GenBank. The closest match for many of them was an environmental sample, accession KM515068, that had been sequenced from an air sample by a lab in Germany. The closest matches for others were three Grimmia pulvinata sequences from California.

Pulling the four relevant sequences from GenBank and adding them to the sequences generated at RBGE showed that there is quite a lot of genetic variation between these accessions. In the phylogram below, specimens identified as “cf pulvinata” were lacking mature sporophytes. When the substrate a plant was growing on is known, it is given (ETICS stands for External Thermal Insulation Composite System). Parsimony bootstrap values are given on a few of the branches; however, the sequences tend to only differ by one or two base pairs and one or two gaps, meaning that statistical support for groupings is not going to be high.
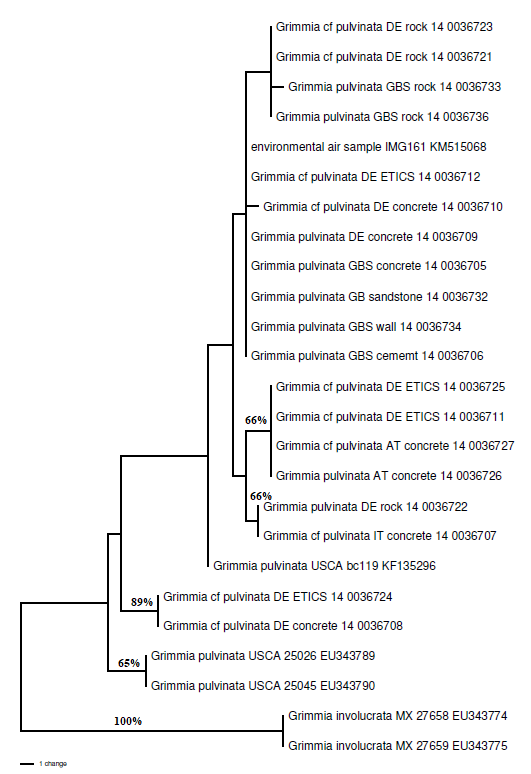
Because ITS2 is a non-coding region of DNA, in addition to variation in nucleotide sequences between accessions (which is visible in the phylogram shown) there are also gaps (insertions and deletions) in the sequences that do not show up in this analysis. For example, the four accessions from Germany and Austria that have 66% parsimony bootstrap support (a statistical support measure for groupings of taxa, obtained through random resampling of proportions of the original DNA sequence data) in the phylogram also share a unique two base pair deletion.
This analysis shows that there are several genetically distinct lineages within the morphological species Grimmia pulvinata – within ten German accessions there are six different DNA sequences, while we have three different DNA sequences within six Scottish accessions.
Intriguingly, there is a small lineage coutaining four accessions, two from Germany and two from Scotland (from the coastal Borders town of St Abbs – or New Asgard, as Avengers fans may better know it, and the Pentland hills near Edinburgh), that were all collected on rocks as opposed to on manmade surfaces.
Without far more thorough sampling, both in terms of accessions of plants and quantities of genetic data, it is only possible to speculate, but it may well be that different lineages within Grimmia pulvinata correlate with some of the distinct substrates that the species is known from.
Even with a plant as common, and as commonly overlooked, as this pollution-tolerant urban bryophyte, there is still genetic diversity to explore and explain.
.
Voucher and collection information for new RBGE Grimmia pulvinata and Grimmia cf pulvinata sequences
| RBGE DNA no. | Voucher | Location | Date | Notes |
| 14-0036705 | Hofbauer 174 | Scotland, Edinburgh, RBGE | 11/09/2014 | concrete wall |
| 14-0036706 | Hofbauer 173 | Scotland, Edinburgh, Ferry Road | 12/09/2014 | cement of stone wall |
| 14-0036707 | Hofbauer 163 | Italy, Triest, near Grotta Gigante | 01/06/2014 | concrete |
| 14-0036708 | Hofbauer 121 | Germany, Valley, Bavaria | 29/08/2014 | no sporophytes; concrete |
| 14-0036709 | Hofbauer 116 | Germany, Valley, Bavaria | 29/08/2014 | concrete |
| 14-0036710 | Hofbauer 094 | Germany, Valley, Bavaria | 29/08/2014 | concrete |
| 14-0036711 | Hofbauer 084 | Germany, Valley, Bavaria | 29/08/2014 | tiny, no sporophytes; ETICS |
| 14-0036712 | Hofbauer 073 | Germany, Valley, Bavaria | 29/08/2014 | tiny, no sporophytes; ETICS |
| 14-0036721 | Hofbauer 110 | Germany, Valley, Bavaria | 29/08/2014 | no sporophytes; rock |
| 14-0036722 | Hofbauer 106 | Germany, Valley, Bavaria | 29/08/2014 | rock |
| 14-0036723 | Hofbauer 103 | Germany, Valley, Bavaria | 29/08/2014 | no sporophytes; rock |
| 14-0036724 | Hofbauer 069 | Germany, Valley, Bavaria | 26/08/2014 | no sporophytes; ETICS |
| 14-0036725 | Hofbauer 062 | Germany, Valley, Bavaria | 26/08/2014 | no sporophytes; ETICS |
| 14-0036726 | Hofbauer 009 | Austria, Kufstein, Tirol | 17/08/2014 | concrete |
| 14-0036727 | Hofbauer 006 | Austria, Kufstein, Tirol | 17/08/2014 | concrete |
| 14-0036732 | Kungu S277 | Scotland, Dumfries & Galloway, Eliock Wood | 24/10/2003 | top of sandstone bridge parapet |
| 14-0036733 | Long 36320 | Scotland, Berwickshire, St. Abbs | 29/09/2006 | rocks |
| 14-0036734 | Long 31186 | Scotland, Lauder Burn | 28/05/2002 | old limy wall |
| 14-0036736 | Long, Chamberlain, Flagmeier 36630 | Scotland, Pentland Hills | 04/04/2007 | rocks |
