Looking at the capture plates from the two DNA extraction protocols that were tested on our QIAcube, it was fairly obvious that a lot more plant fragments and pigment were transferred over using the QIAamp kit. I started wondering what on earth I was doing this for – after all, why compare a kit that’s designed for plants with one that isn’t (and where the protocol keeps referring to blood or to mouse tails…).
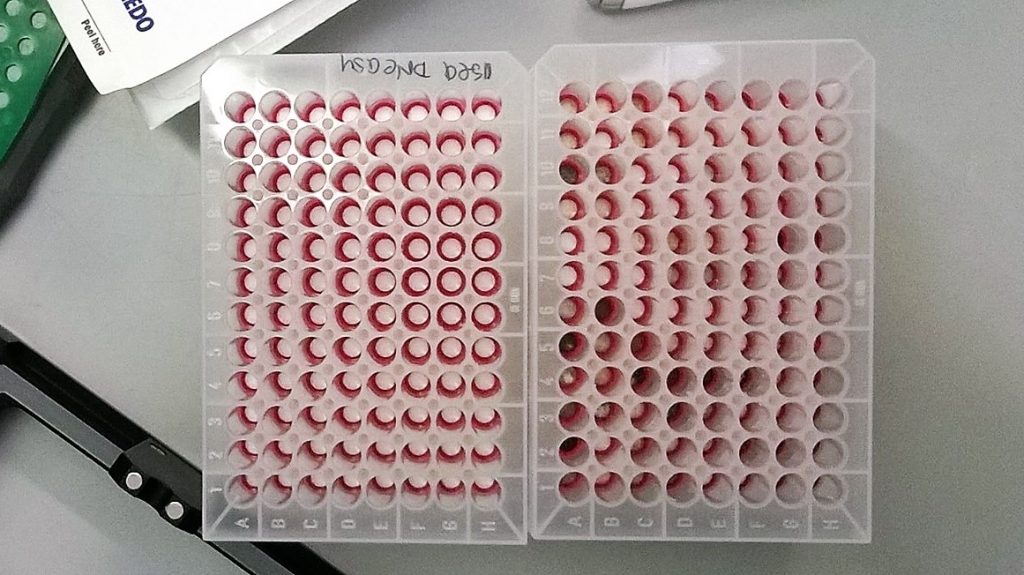
Used capture plates from the DNeasy Plant 96 (lhs) and the QIAamp 96 QIAcube HT (rhs) extraction kits
However, having done the two plates of extractions, there was nothing to be lost by running them out on agarose gels (100 μl 2% agarose TBE gels with 5 μl SYBRsafe and 3.5 μl 1kb+ ladder on alternate rows; loading 2.5 μl loading buffer and 5 μl of each DNA extraction). I did cut corners a little by using our multichannel gel set-up, which doesn’t let the samples run as far as a manually loaded gel, but did let me get them both run in a short afternoon!
And so, here are the gels….
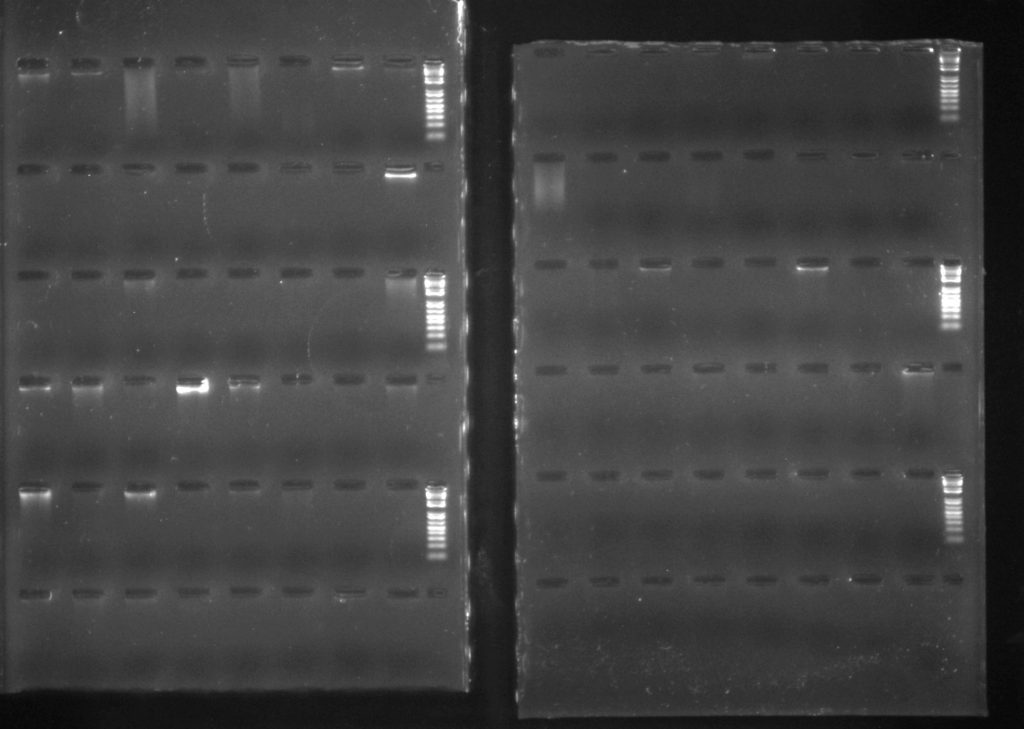
DNeasy Plant 96 extractions: Samples: 1A-1H: Conifers (silica); 2A-2G: Gingers (herbarium); 2H-6B: Sapotaceae (silica); 6C-7H: liverworts (silica); 8A-8F: Bamboos (herbarium); 8G-8H: Begonia (silica); 9A-10H: lichens (herbarium); 11A-11H: bryophytes (herbarium); 12A-12H: Begonia (silica)
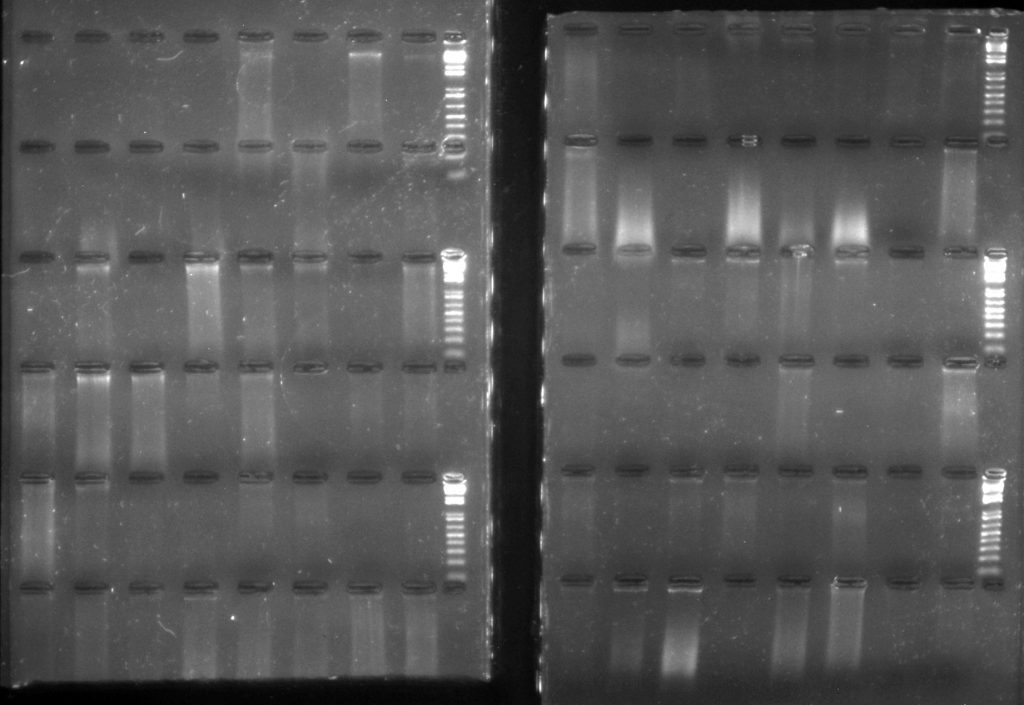
QIAamp 96 DNA QIAcube HT kit extractions:Samples: 1A-1H: Conifers (silica); 2A-2G: Gingers (herbarium); 2H-6B: Sapotaceae (silica); 6C-7H: liverworts (silica); 8A-8F: Bamboos (herbarium); 8G-8H: Begonia (silica); 9A-10H: lichens (herbarium); 11A-11H: bryophytes (herbarium); 12A-12H: Begonia (silica)
The QIAamp extractions are a lot more smeary, which is curious: it’s possible that the kit doesn’t do as good a job at removing some of the secondary chemicals which could be co-extracting with the DNA. Some of the DNA elutions are straw-coloured, unlike the colourless elutions from the Plant DNeasy kits.
This is only the first step in comparing these two QIAcube extraction kits: I will take Qubit fluorometer readings to look at concentration, and NanoVue spectrophotometry readings to get an idea about purity. I’ll also run some of the samples on our TapeStation, to get a better idea about the distribution of recovered DNA fragment sizes.
But for now it’s safe to say, having run these quick gels, I’m rather less ready to dismiss the QIAamp kit!
————————————————————————————————
Related Posts
————————————————————————————————
